Martin, thanks very much for the build instructions.
Two further comments:
Line 2500 of lcmodel.f, add a line reading go to 200 to bypass the license check altogether, and obviate the need for adding the master key to your .control files.
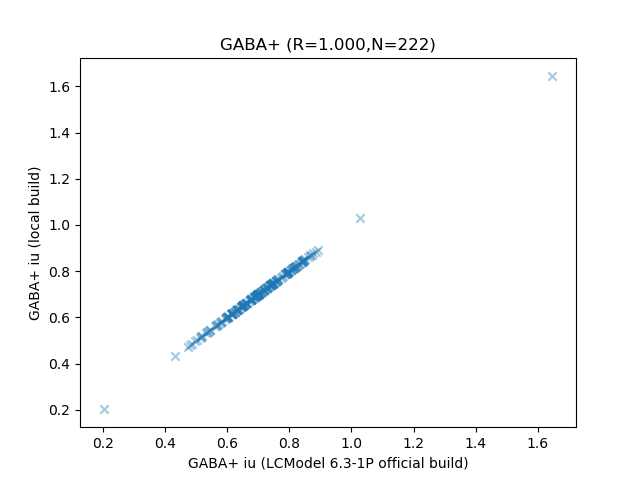
Regarding numeric differences, I just ran the full public Big GABA dataset through a local build (Debian 9.1, gfortran 6.3.0-18+deb9u1), I’d say it agrees pretty well in this instance ![]() Of course this may well differ according to library versions…
Of course this may well differ according to library versions…
(N=222, total processing time 8.65 seconds with a bit of parallelisation: it’s fast)