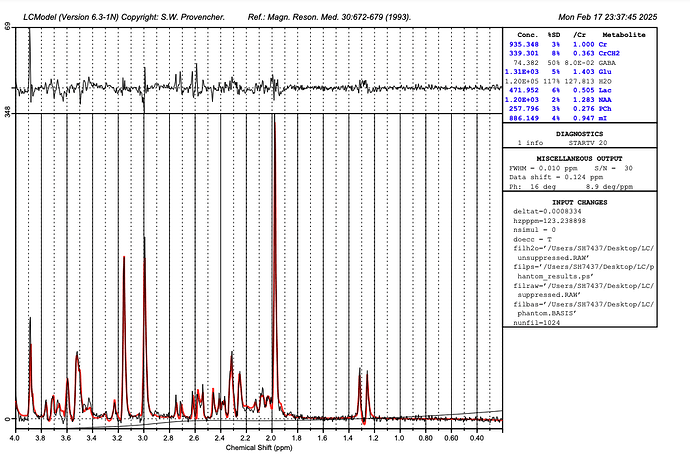
But should not I also need to add my unsuppressed file as well?
This looks much better.
You can add the water-unsuppressed file (add it as filh2o to the control file), yes, and also set the doecc = T flag, which might help with the lineshape a little bit.
Deactivate all the MMs and lipids (nsimul = 0) as well. I’m not sure but I think MRSCloud also allows free choline to be simulated, which is probably a better spin system for the choline chloride in the phantom than the PCh you’re using.
Thanks
I did, they improved the %SD by 1%
Does the residual look better?
I’m looking to expand my current MRS skills aiming at quantifying GABA using MEGA-PRESS, sequence. I would greatly appreciate your opinion about using MRScloud for simulating basis files is a valid option? Also, any suggestions on getting the most out of our SPECTRE phantom (from Gold Standards) for validation or quality control would be really helpful, should I expect to get exact measure of GABA as in the phantom’s data sheet.
Thanks
-
I’m biased because I contributed to MRSCloud, but I think it produces good results for standard product sequences with minimal user input and expertise required (which was always the point). If you need to have exact control over the timing (which can fluctuate ever so slightly depending on hardware configurations), or if you are using a rare or custom sequence, you’ll want to look into different options, but this is definitely a more advanced skill (and you’ll need access to often proprietary information such as sequence timings, waveforms etc.)
-
We usually only use phantoms to make sure that our newly developed sequences behave in the way they should. For quantitative validation or anything, they’re pointless IMO - as described above, quantification requires detailed knowledge of relaxation behavior, which is complicated to measure. (This is even more true for MEGA-editing, which operates at longer TE, making the signals even heavier T2-weighted, and the T2 of coupled signals is even harder to measure).
-
Generally, I would highly recommend that you start gaining experience with a simple experiment (un-edited short-TE data) in human participants (acquiring good-quality data and complete data processing) before you turn towards edited experiments (they’re much more demanding on data acquisition and analysis).
Thank you so much for your feedback and help! I greatly appreciate it.