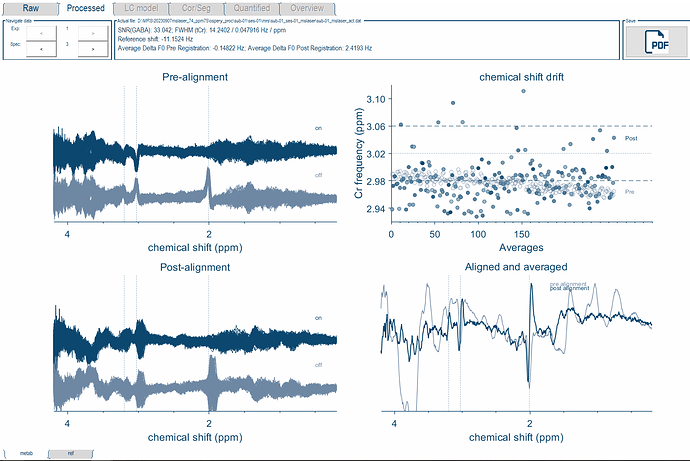
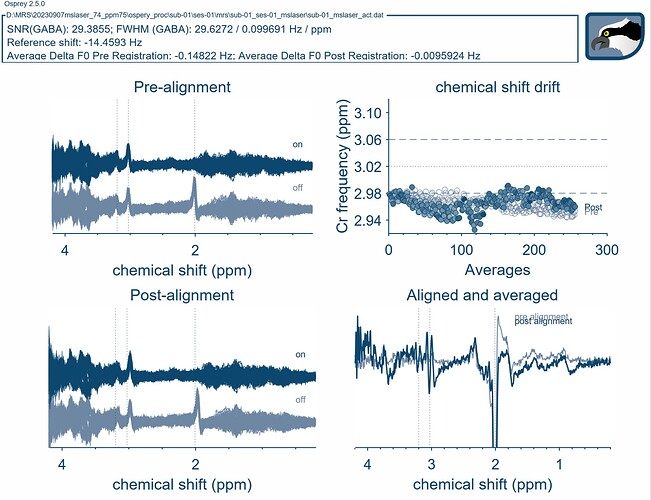
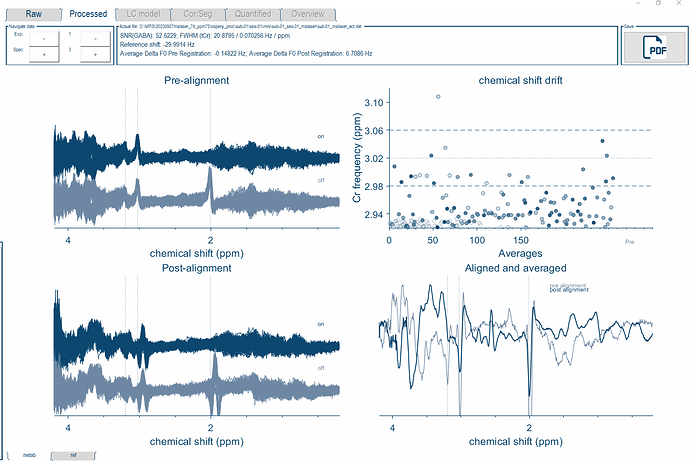
Hi Helge,
Thanks so much for your reply. I will share our data via email.
As for the LCModel basis set, I just updated my folders to most recent update in the develop branch. I was able to import the basis file into the Osprey format using the io_LCMBasis function. I used this new file for the fitting and it worked! I just had to add an if statement for the Siemens case in osp_fitInitialise, so that it would load the correct basis set.
I will have to look into the results more closely, but this is a great start.
I did get an error towards the end when calculating the correlations between metabolites and SNR/LW in the overview tab:
Unrecognized function or variable ‘lgamma’.
Error in betapdf (line 60)
+ lgamma (a + b) - lgamma (a) - lgamma (b));
Error in betainv (line 87)
h = (betacdf (y_old, a, b) - x) ./ betapdf (y_old, a, b);
Error in finv (line 58)
inv(k) = ((1 ./ betainv (1 - x(k), n/2, m/2) - 1) * n / m);
Error in regression_line_ci (line 59)
Yoff = (2*finv(1-alpha,2,N-2)*SE_Y).^0.5;
Error in osp_plotScatter (line 152)
[~, ~,~] = regression_line_ci(0.05,b,x_tmp,y_tmp,cb(g,:));
Error in osp_iniOverviewWindow (line 293)
[temp] = osp_plotScatter(MRSCont, gui.quant.Names.Model{gui.quant.Selected.Model},
gui.quant.Names.Quants{gui.quant.Selected.Quant},MRSCont.quantify.metabs{gui.overview.Selected.Metab},MRSCont.QM.SNR.A’,gui.overview.Names.QM{gui.overview.Selected.Corr});
Error in OspreyGUI (line 408)
osp_iniOverviewWindow(gui);
Any ideas as to why this happened? I don’t remember running into this issue before.
Thanks again!
Best,
Humberto
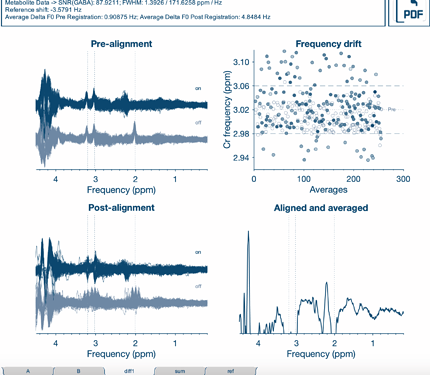
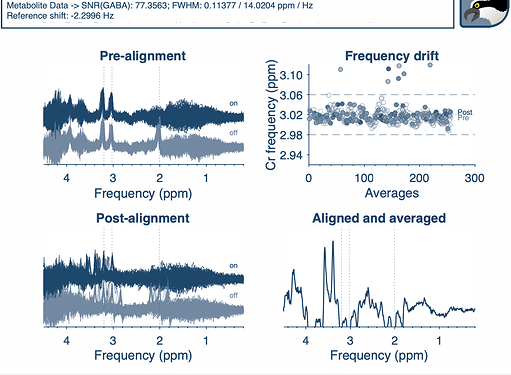

 ).If it is of enough interest to you, I would be happy to provide different test data for implementing this and possibly give a manuscript of its own? Not sure if this is doable (I’m still fairly new to all of this
).If it is of enough interest to you, I would be happy to provide different test data for implementing this and possibly give a manuscript of its own? Not sure if this is doable (I’m still fairly new to all of this  ).
).