Hi! I’m new to MRS Hub and I’ve just started doing research in Magnetic resonance spectroscopic imaging (MRSI). I’m currently working with SPAR and SDAT files by using Python and I’m facing some problems to deal with the spectra’s edges.
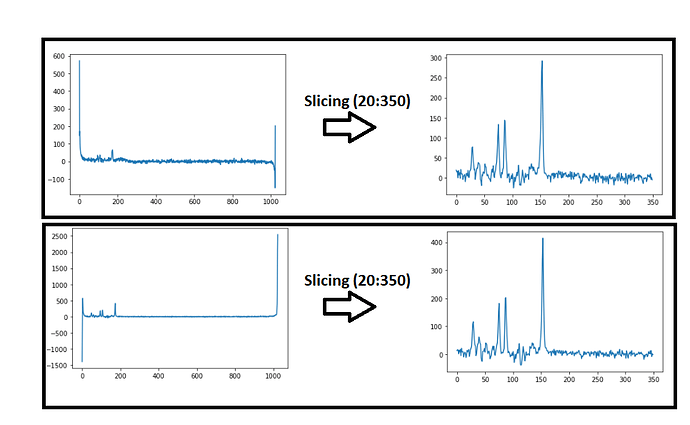
At the edges of my spectra signals, their amplitudes have really high values, but I´m almost confident that this is not related to a biological metabolite nor water influence. I’d like to know if this configurates an artefact problem intrisic to the spectras from SPAR/SDAT files.
My spectra signals have 1024 points. To get a better spectra visualization, I´m doing a slice. It can be seen in the picture that I´am attaching in this post.
I’d like to know what is the best data processing technique to deal with this. Also, I’d like to know why this edge occurs.
Thank you for the help and attention.