- I changed, with the previous version of Osprey, the range of the HSVD water filter from [4.6 4.8] to [4.5 4.9]. The fit is much better.
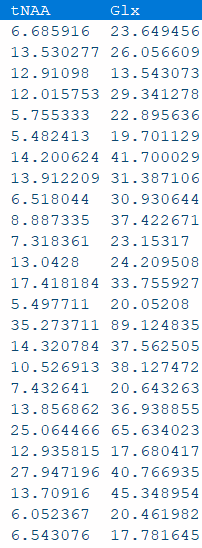
Nevertheless, I found strange data (figure below). As far as I know, the [tNAA] > [Glx], but I did not find this in my data. Do you have some idea to explain that?
-
Where is it possible to find the quality-control data from each spectra, for example in the overview section?
-
With the last release of Osprey, I obtain an bug directly after the OspreyProcess function:
Undefined function or variable ‘osp_lcmcontrol_params’.
Error in osp_saveLCM (line 53)
LCMparam = osp_lcmcontrol_params(MRSCont.flags.isMEGA);Error in OspreyProcess (line 99)
[MRSCont] = osp_saveLCM(MRSCont);
Thank you and I appreciate any reply.
Frederic