Hi everyone!
I want to call the conversion functions from spec2nii through a python script instead of using the command line.
I’m studying and trying to understand the source code from spec2nii GitHub - wtclarke/spec2nii: Multi-format in vivo MR spectroscopy conversion to NIFTI but it has been really hard for me to figure out how I would be able to do it.
Can you help me please to manage this so I can call spec2nii functions in a python script?
Thank you
Hi @Gabriel_Dias ,
You can probably call the functions stored in specific modules for different file formats. For example, if you wanted to manually handle Philips SPAR/SDAT files, you can have a look a the main spec2nii.py file and the respective call for that file type: spec2nii/spec2nii.py at master · wtclarke/spec2nii · GitHub
You could run the conversion as following:
from spec2nii.philips import read_sdat_spar_pair
from pathlib import Path
sdat = Path('path/to/my/data.SDAT')
spar = Path('path/to/my/data.SPAR')
converted_data = read_sdat_spar_pair(sdat, spar, None, ["DIM_DYN", None, None])
converted_data.save('output/location/my/data.nii.gz')
Hope that helps.
1 Like
Hi @wclarke ,
That’s exactly what I want to do. I’m working with Philips SPAR/SDAT files. I tried to use the code in your answer but I got the following error:
“AttributeError: module ‘spec2nii.nifti_mrs’ has no attribute 'NIfTI_MRS”
Do you know what is wrong?
Thank you very much for your help.
Could you post the full traceback?
What version of spec2nii are you using? If you have conda run conda list and it will list a version.
Will
I’m using the version 0.4.8 of the spec2nii.
The full traceback:
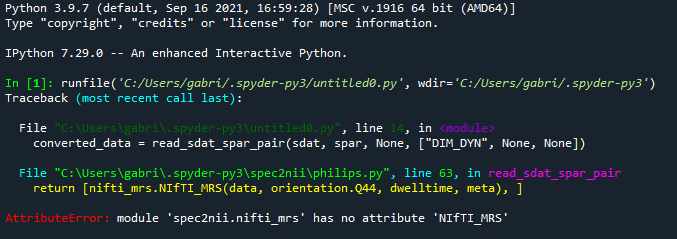
Thank you the attention @wclarke
Hi @Gabriel_Dias ,
I think you’ve definitely found a (sort of) bug, a result of the functions not being designed to be called separately from the spec2nii.py code.
The nifti_mrs.NIfTI_MRS class has been loaded/imported in the spec2nii function by this code nifti_mrs.NIfTI_MRS = nifti_mrs.get_mrs_class(nifti=2). This is a part that allows nifti-1 or nifti-2 to be selected at a global level. I think that if you run that line in addition to the code that I posted above it should work.
from spec2nii import nifti_mrs
nifti_mrs.NIfTI_MRS = nifti_mrs.get_mrs_class(nifti=2)
from spec2nii.philips import read_sdat_spar_pair
from pathlib import Path
sdat = Path('path/to/my/data.SDAT')
spar = Path('path/to/my/data.SPAR')
converted_data = read_sdat_spar_pair(sdat, spar, None, ["DIM_DYN", None, None])
converted_data.save('output/location/my/data.nii.gz')
1 Like
Hi @wclarke
It worked now! But I had to do one more alteration.
In the last line of the code from your last answer I got the following error: "AttributeError: ‘list’ object has no attribute ‘save’ "
Then I changed this last line to:
converted_data[0].save('output/location/my/data.nii.gz')
And it has worked.
Could you tell me please the difference between nifti-1 and nifti-2?
Thank you very much for everything 
Nifti-2 was a refinement of the nifti-1 header. See NIfTI-2 Data Format — Neuroimaging Informatics Technology Initiative.
There’s the option in the code because some tools might only handle one or the other.
1 Like
Thank you very much @wclarke!
I hope that other people can benefit from this topic as well.
