Hi,
I recently started using FSL-MRS for analyzing MEGA-PRESS data. I was able to fit the spectra successfully, but noticed that the quantification output was missing individual tissue volumes. Additionally, the tissue-water density values were the same across all subjects: GM = 0.78, WM = 0.65, and CSF = 0.97 (g/cm³).
After discussing this with my team, we agreed that it’s essential to use accurate, subject-specific tissue volume information. I’m now trying to use svs_segment to extract these values.
For inputs:
- T1w: I’m using either
norm.niifrom the FreeSurfer pipeline orT1_biascorr.niifrom the FSLfsl_anatpipeline. - Mask: This is where I’m encountering an issue.
Based on my acquisition protocol, it doesn’t seem like an explicit mask was provided. The sequence looks like this:
01-localizer_32Channel02-acq-MPRage_T1W(my anatomical reference)03-recon_ax04-recon_cor05-eja_svs_mpress_lw06-eja_svs_mpress_lw07-VOI_precuneus08-eja_svs_mpress_On(MRS data – transients)09-eja_svs_mpress_On_RES(individual pulses?)
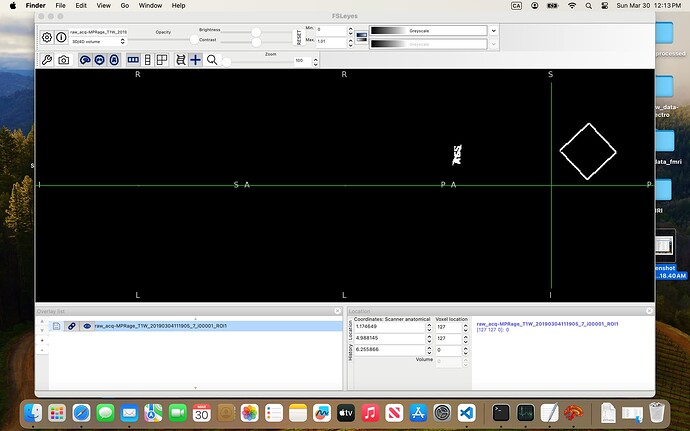
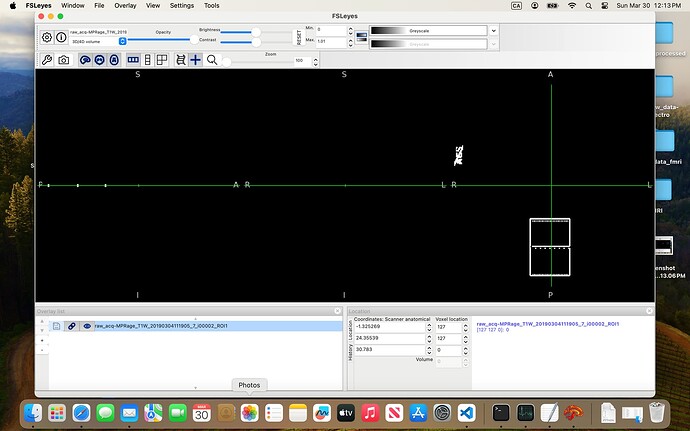
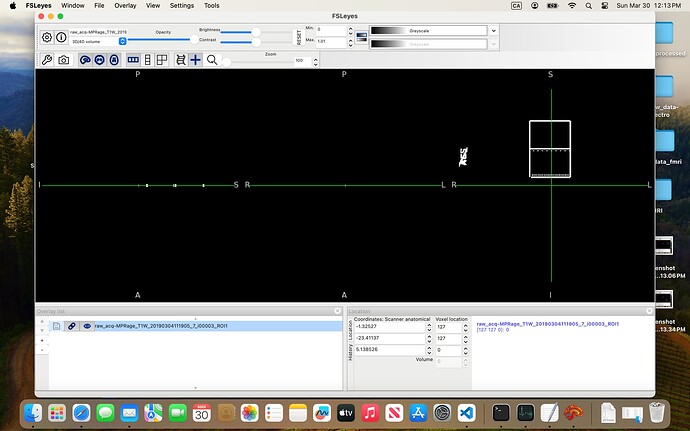
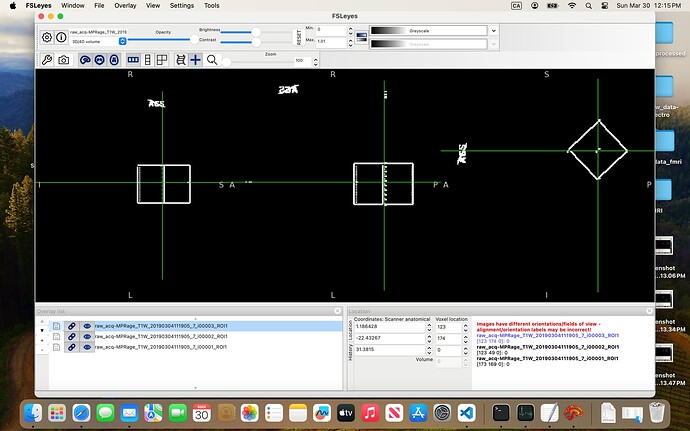
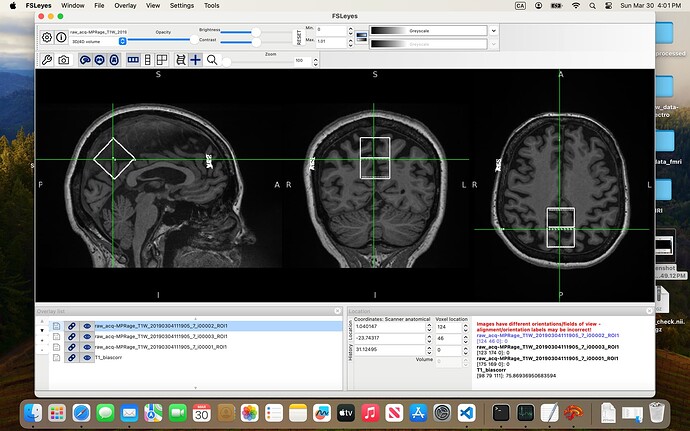
In the 07-VOI_precuneus folder, I have three DICOM files which, when converted to NIfTI, generate multiple files. When viewed in FSLeyes (see below), three of them — i00001_ROI1, i00002_ROI1, and i00003_ROI1 — clearly show box-like regions, presumably corresponding to the VOI. The rest seem to be duplicates or represent the slice on which the box was positioned.
I’ve been trying to combine these three ROIs and register them to the anatomical image so I can use the result as the mask input to svs_segment, but I keep running into errors.
Am I on the right track? And would you be able to guide me on how to properly create a usable mask from these files?
Thank you very much in advance!
Best regards,
Arsenii