Hi all,
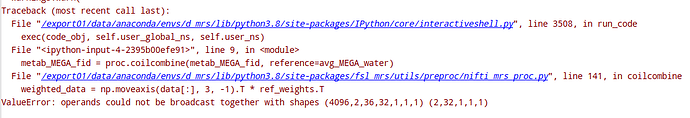
I’m trying to pre-process our MEGA-sLASER data, acquired at 7T using FSL-MRS. Data dimensions are in (x,y,z, n_points, n_coil, n_averages, n_editing) format. However, I’m running into the following error when running coil combination:
It seems the function doesn’t handle the editing dimension well. I tried tweaking the FSL-MRS coilcombine() function, but I couldn’t make it work. How can I fix this problem?
I really appreciate your help and thanks so much in advance!
Cheers,
Hande
UPDATE: Fixed the error by adding 2 more dimensions to ref_weights using numpy.expand_dims(). I aimed to match the dimensions of data after moveaxis() operation. I also tweaked the functions to get sepearate coil noise covariance and weights for edit on and edit off datasets.
Just started pat leave. Will try to get back to you soon.
I see I managed to undo some of my work to implement more helpful error reporting when improving things for MRSI processing.
What’s the shape of the two imputs to proc.coilcombine? I.e. metab_MEGAfid.shape and avg_MEGA_water?
I also tweaked the functions to get sepearate coil noise covariance and weights for edit on and edit off datasets.
This shouldn’t really be necessary because you expect the same coil combination solution for each editing case. Have you collapsed the editing dimension in your reference data?
Hi @wclarke!
Thank you for getting back to me during your pat leave. Hope you and your family are well!
The shape of the inputs: metab_MEGA_fid: 1,1,14096,32,36,2, water_MEGA_fid: 1,1,1,32,2.The 2 at the end represent “EDIT ON” and “EDIT OFF”. The data is single voxel.
This shouldn’t really be necessary because you expect the same coil combination solution for each editing case.
I thought we needed different sets of weights for edit on and off. Can you explain why we expect the same coil combination solution for both editing cases?
To implement different sets of covariance matrices and weights for each editing case, I added an extra for loop to iterate over the editing case dimension. Then I added 2 extra dimensions, equal to 1, to ref_weights array with np.expand_dims() to match the dimensions of ref_weights and metab_MEGA_fid.
I really appreciate your help, thanks again so much.
Best,
Hande
Hi @handehio,
Yes, so the solution to this is to average the edit dimension in the water_MEGA_fid case before using it as the water reference for coil combination. Something like:
averaged_water = proc.average(water_MEGA_fid, dim='DIM_EDIT')
combined = proc.coilcombine(metab_MEGA_fid, reference=averaged_water)
The combination should be the same as the data was acquired from the same voxel and same coils. The weighting is a function of the voxel to coil element distance (and path through the tissue), and the covariance is a function of the position of the subject relative to the coils and each other. Neither of thes things change between the (usually) interleaved edit on and off conditions.
Will
1 Like
Awesome thank you so much!