This suggests that modeling for those failed completely. Can you post what the spectra look like? If loading/processing takes forever, chances are that data quality is so bad that they need to be removed.
Apologies for the delay! Images of the processed data are below.
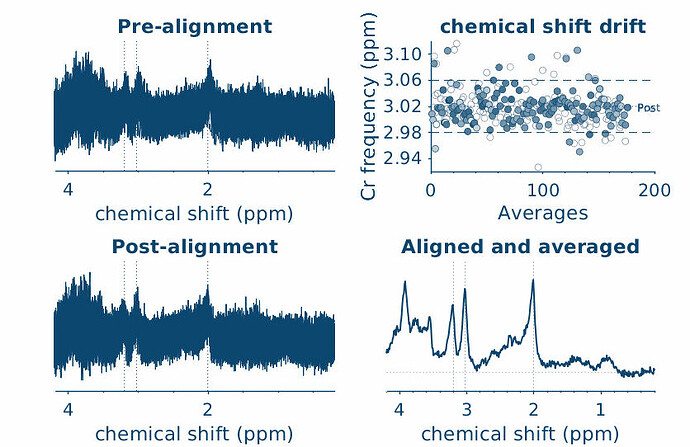
sub-1601_3
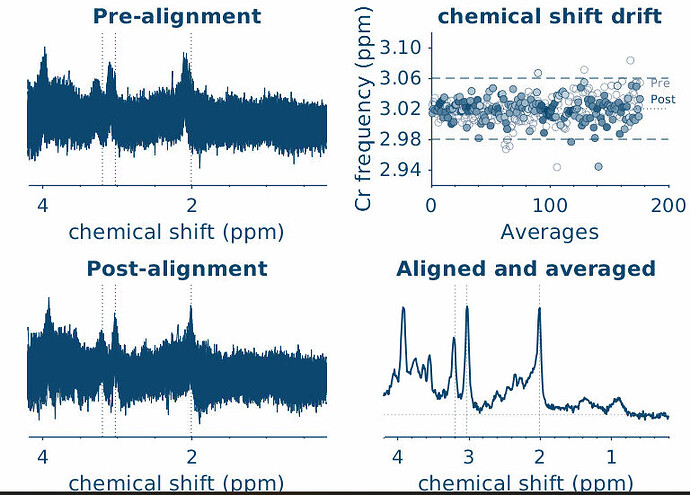
sub-1602_1
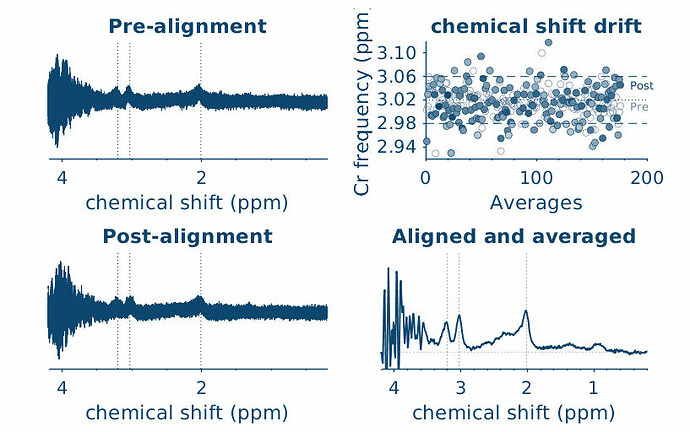
sub-1606_1
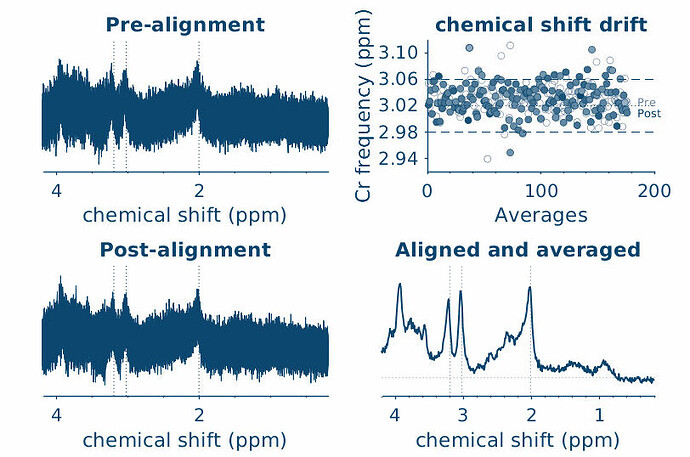
sub-1608_1
I’m thinking the spectra look reasonably decent, except for perhaps 1606_1. Is there anything that I can do to salvage these data for analysis?
Many thanks!
–Kimberly
I agree, they don’t look bad at all, and even the third one should get fitted. Can you check the LCMoutput folder in your specified output folder if there are .ps or .print files for the failed datasets?
They all for look the same as below…
sub-1601_3
See any clues as to what I may be able to do?
Many thanks!
–Kimberly
So the MYDATA 10 error, according to the LCModel manual, reports on this:
(FATAL) Error trying to read time data in FILH2O.
Hints: In FILH2O, check that the time data correspond to FMTDAT. Be sure that FILH2O corresponds exactly to FILRAW, except for water suppression, especially for multi-voxel data sets.
This means LCModel cannot read the water reference data. Do you see anything unusual about the water reference data when you look at them in Osprey? Can you confirm that they were acquired with the exact same parameters (TE, number of points, spectral width) as their metabolite counterparts?
You can compare the headers of the .RAW files that Osprey writes out to the LCModelFiles directory (with metabs and ref subfolders) in your output folder - just open them in a text editor and compare the first couple of lines.
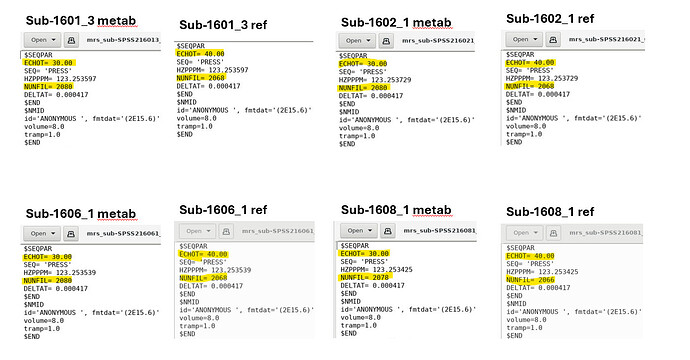
Unfortunately, there do appear to be some discrepancies between the metab and ref files generated by Osprey in the LCModelFiles directory…
Specifically, the ECHOT and NUNFIL values are different within-subjects. The HZPPM value seems to vary across participants but is the same within-subjects. I do not see any reference to FILH20…
Is there anything I can do to assist LCModel in reading the water reference data?
Many thanks!
–Kimberly
Yeah, so, the water references for these datasets were acquired at different echo times and numbers of data points than the metabolite scans. That’s a problem. I suppose this didn’t happen for the datasets that don’t throw this error? (you can also compare the headers for those)
(FILH2O is just the field in the control file that points to the path of the water reference file - it’s not expected to be in here)
Correct, this error did not occur for the other datasets that worked, I checked a few of the .RAW files and the values are all the same…
Is there anything I can do to salvage these 4 datasets?
You guidance and expertise and greatly appreciated!
Much thanks!!
–Kimberly
Water-scaled metabolite ratios will be difficult to be compared between these 4 datasets and the others (because your reference has a different TE). But you can still include them in an analysis of tCr ratios (which don’t need a water reference). You’d just need to run these 4 in a separate Osprey job that doesn’t use the water reference files.
Amazing, thank you!!! I will give it a shot!