hello everyone,
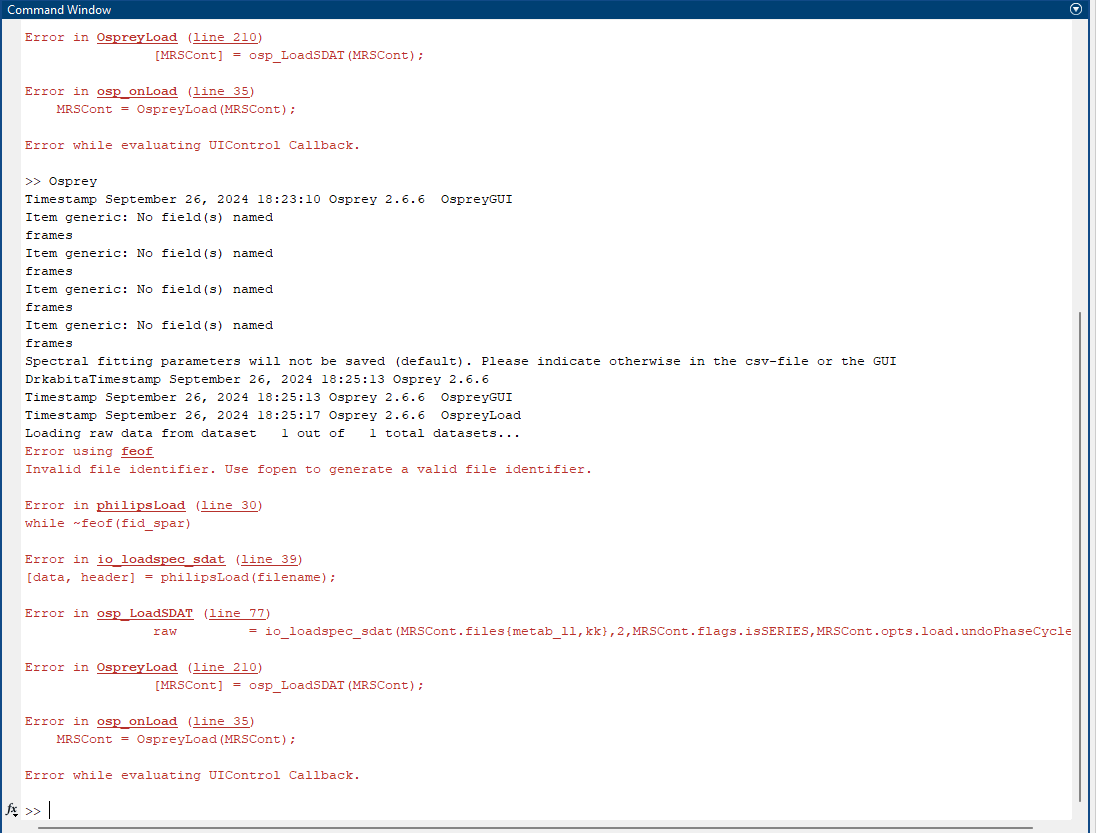
i am having problem in loading job for megapress in osprey. the error message is attached below. please help
The ‘invalid file identifier’ error suggests that your job file is pointing to a file that doesn’t exist on your drive. Please check spellings etc.
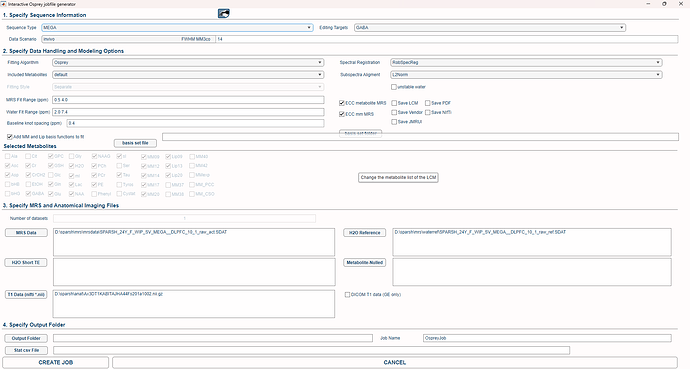
Hi, I am new to both MRS and osprey. this is my create job window for megapress. the basis set file is otherwise empty for press. do i need to upload or select any basis set file for megapress?
No, Osprey comes with basis sets for some of the more popular MEGA-PRESS sequences and should be able to find it automatically.
Thank you for your time. I don’t notice any apparent mistakes in the file paths, spelling, or options selected. Could you kindly check if there’s anything wrong that stands out in the configuration? I’ve attached a screenshot of the jobfile generator in my previous message.
Additionally, could this error be due to an issue with the data files themselves? I’m using Philips SDAT/SPAR files
I’d appreciate any guidance you can provide.
Thank you!
Do the SPAR files have the exact same filenames as the respective SDAT files?
Are they in the exact same folder?
Can you post the contents of the job file you generated?
Hi @Tanmay
I got the same error yesterday. I too have data from the Philips scanner (SDAT & SPAR files), but mine was PRESS sequence. One of the subjects had a corrupted SPAR file, when I excluded this specific subject, the data load worked for me.
In the next screenshot, I see you have only one subject’s files loaded. Could you try loading a different subject’s files? May help!
Best,
Anushree Bose
Thank you so much for your help. I think there was an issue with that SPAR file and after i tried with a different dataset it worked.
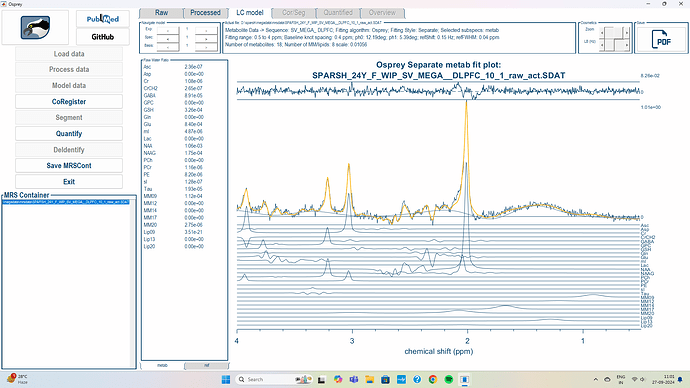
However, I’m still facing a problem. The spectrum doesn’t look as expected, the NAA peak isn’t inverted as it’s supposed to be. What could be the reason?
I’ve attached a screenshot for your reference
Please let me know if you need more information to better understand the issue.
I can share my dataset if you could please look and help identify if there’s an issue with the acquisition or processing.
Thank you again for your support!
Thank you Anushree, it was indeed an issue with that SPAR file and after i tried with a different dataset it worked.
Thank you for reaching out