Great 1/3 fixed!
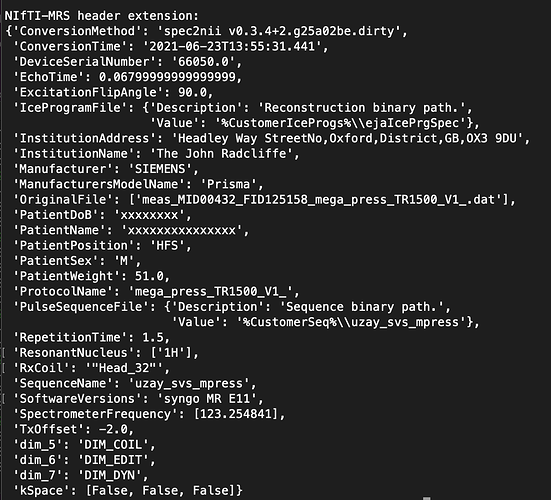
spec2nii dump myfile.nii.gz will print to stdout the Nibabel formatted NIfTI header and the NIfTI-MRS header extension (where all the MRS specific metadata is stored). The latter might look something like this
There are two required keys in this which are needed ‘SpectrometerFrequency’ and ‘ResonantNucleus’. In my example they are [123.25…] and [‘1H’]. Could you run the command on your data and post a picture here of what the ‘SpectrometerFrequency’ value is?
Looking closely at the error I think you might also have an old version of FSL-MRS (maybe one which comes with the main FSL installation). If you run fsl_mrs --version it should give you the version string. The most recent version is 1.1.2. I think you might have a version that was pre-NIfTI-MRS becoming the main file type (the problem with developing the data standard after the analysis program).
You can install the new version by following the instructions here.
For svs_segment what it does under the hood is:
- Runs fsl_anat on the T1. fsl_anat does: fslreorient2std, robustfov, FAST bias field correction, registration to standard space (linear and non-linear) [FLIRT and FNIRT], brain-extraction [FNIRT-based or BET] and finally tissue-type segmentation [FAST].
- Creates some 2x2x2 mock data with the same orientation affine as the svs voxel. Effectively a low resolution mask.
- Uses FLIRT to register that mock data to the T1 to create a mask for the svs voxel in T1 space.
- Masks and runs fslstats (mean) on the
pve_noutputs of FAST.
I think the messages you get about linear registration are coming from some of the steps run in fsl_anat. These steps aren’t used in the calculation of the FAST outputs. To check this you can spoof the fsl_anat step, replacing it with your own FAST output. Create a folder my_spoofed.anat and pass it to svs_segment as the -a option (also don’t include the -t option). In the folder you need a T1 called 'T1_biascorr.nii.gz' and the output of FAST 'T1_fast_pve_0.nii.gz', 'T1_fast_pve_1.nii.gz' and 'T1_fast_pve_2.nii.gz'. This will then not run the fsl_anat step and substitute your data into the calculation.
Of course you can also check what everything looks like in fsleyes. You can load the FAST results from both pipelines and compare. You can also load the mask created by svs_segment and verify that it matches the original svs voxel closely.