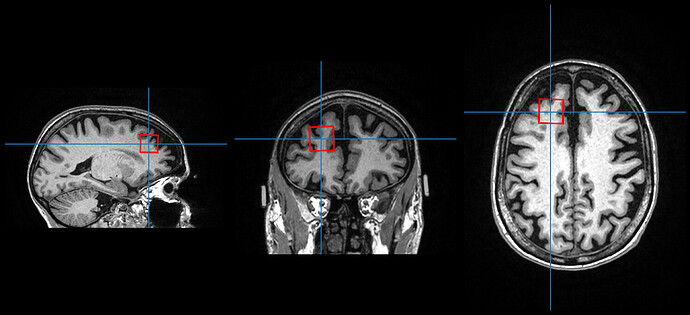
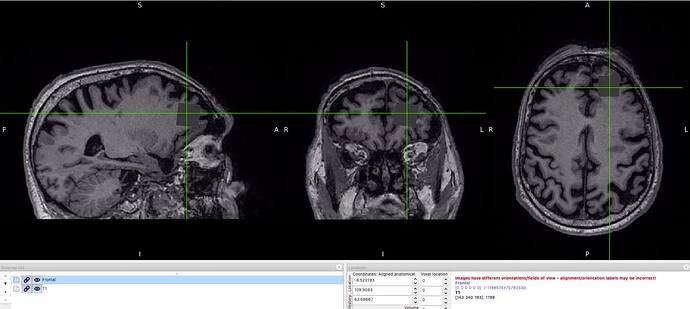
Hi William,
No worries at all, and congratulations! I ran fslhd on both and I’m pasting the data for T1 and SVS below:
filename Anatomical/T1.nii.gz
sizeof_hdr 348
data_type INT16
dim0 3
dim1 372
dim2 512
dim3 324
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 4
nbyper 2
bitpix 16
pixdim0 1.000000
pixdim1 0.500000
pixdim2 0.500000
pixdim3 0.500000
pixdim4 2.390720
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 0.499025 -0.031155 -0.001820 -87.371529
qto_xyz:2 0.031202 0.498644 0.019510 -51.212696
qto_xyz:3 0.000599 -0.019585 0.499616 -30.942078
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Left-to-Right
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 0.499023 -0.031155 -0.001820 -87.371529
sto_xyz:2 0.031202 0.498644 0.019510 -51.212677
sto_xyz:3 0.000599 -0.019585 0.499616 -30.942078
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Left-to-Right
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip FSL5.0
filename MRS/proc/Frontal.nii.gz
sizeof_hdr 540
data_type COMPLEX64
dim0 6
dim1 1
dim2 1
dim3 1
dim4 4096
dim5 12
dim6 12
dim7 1
vox_units mm
time_units s
datatype 32
nbyper 8
bitpix 64
pixdim0 -1.000000
pixdim1 20.000000
pixdim2 20.000000
pixdim3 20.000000
pixdim4 0.000200
pixdim5 1.000000
pixdim6 1.000000
pixdim7 1.000000
vox_offset 1344
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name mrs_v0_8
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Aligned Anat
qform_code 2
qto_xyz:1 -20.000000 0.000000 -0.000000 -20.909908
qto_xyz:2 0.000000 -20.000000 -0.000000 117.590614
qto_xyz:3 0.000000 0.000000 -20.000000 66.839722
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Anterior-to-Posterior
qform_zorient Superior-to-Inferior
sform_name Aligned Anat
sform_code 2
sto_xyz:1 -20.000000 0.000000 0.000000 -20.909908
sto_xyz:2 0.000000 -20.000000 0.000000 117.590614
sto_xyz:3 0.000000 0.000000 -20.000000 66.839722
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Anterior-to-Posterior
sform_zorient Superior-to-Inferior
file_type NIFTI-2+
file_code 2