Hi, i have a question, I want to import metabolite “carnosine” in Osprey and I don’t understand how… I know in the guide osprey there are a command fit_makeBasis but I don’t know how use it…
I work with Siemens 3T…

Hi Justine,
We’ll need a bit more information here.
Import from where or which format? Do you have a complete basis set that includes carnosine? If yes, in what format? Or do you just have a simulated carnosine signal that you want to add into an existing basis set? (Also, is it carnosine or homocarnosine?)
Thanks
Georg
Hi,
Its for import from fid-a which is in .mat format we would like to modify it with the characteristics of the machine (therefore make a simulation) then put it in osprey under .rda in order to use it commonly in osprey with the graphical interface
Thanks
Justine
Hi Justine,
Apologies for letting you wait - I’ve been traveling to the ISMRM which is happening this week!
So, we’ll still need to clarify a little what you have and what you need:
- Do you have an existing basis set or do you want to use one that Osprey has and just add a metabolite (what echo time? which sequence? which vendor?)?
- Are you talking about carnosine or homocarnosine? We have the spin system definitions for the latter, but not for the former, we’d have to do a bit of research.
Thanks,
Georg
Hi Georg no problem !
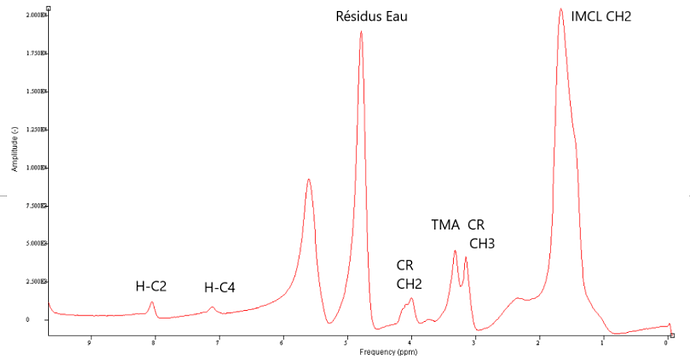
Here is a muscle spectrum
We are interested in the carnosine peak of C2 and C4
Hi Justine,
LCModel has very specific settings for muscle spectra, and it uses internally simulated signals instead of externally simulated basis sets. We might have to add a few edits to the way that Osprey handles this kind of data, because so far we have only really done brain spectra. Mainly this will include some tailoring of the control file that will need to be fed into LCModel. Have you consulted the LCModel manual already?
Thanks,
Georg
Hello guys @admin @justinejetu ,
I’m a new user off Osprey and I also want to import “L-Carnosine” in osprey to study muscle. Is there any edits in Osprey as you mentioned in your previous messages or I have to go through LCModel to add it.
By the way, I would like to thank you for the course online, it help me a lot to understand how to use software !!
Thanks,
Arthur
Hi Arthur,
Thanks for your kind words. I’m not really sure - the default LCModel muscle analysis (fig-muscle) doesn’t seem to include it, can you point me to a reference where this has been done? I’m not too familiar with muscle MRS.
Thanks,
Georg
Thank you for you’re answer. The study is based on this publication : https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/nbm.4266
I would like to add the two frequencies that are shown in the publication above
Thanks,
Arthur
Do you have an example dataset that you can post a screenshot of here? If I understand correctly, the carnosine signals are around 8 ppm. You’d have to create a template LCModel control file that specifies to extend the fit window (ppmst = 8.5 or something) and then add two simulated resonances following Section 11.7 in the LCModel manual.
It’s going to be challenging to do the analysis as you want it in Osprey as is today, mostly because it isn’t currently very well geared towards muscle MRS. We might be able to accommodate that but that would require some work/time. Have you tried processing your data through Osprey at all (before modeling)?