Hi!
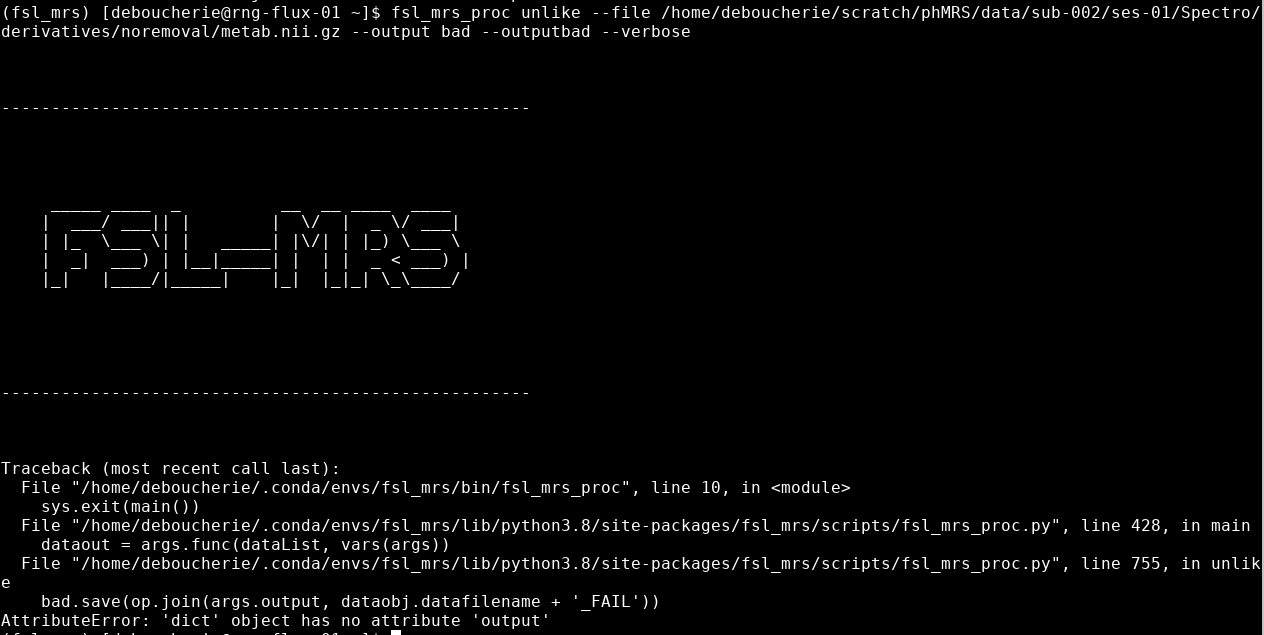
I’m currently analysing my fMRS data using FSL-MRS. For the analysis, I would like to create an output of the bad averages using fsl_mrs_proc unlike, which I can subsequently use to create a subject-specific design matrix (similar to fMRI analysis). However, when trying to run the following command (removed the overall folder structure to increase readability):
fsl_mrs_proc unlike --file ~/fMRS_metab.nii.gz --outputbad --output ~/derivatives/badaverages --generateReport --verbose
I receive this error:
Traceback (most recent call last):
File “/home/deboucherie/.conda/envs/fsl_mrs/bin/fsl_mrs_proc”, line 10, in
sys.exit(main())
File “/home/deboucherie/.conda/envs/fsl_mrs/lib/python3.8/site-packages/fsl_mrs/scripts/fsl_mrs_proc.py”, line 428, in main
dataout = args.func(dataList, vars(args))
File “/home/deboucherie/.conda/envs/fsl_mrs/lib/python3.8/site-packages/fsl_mrs/scripts/fsl_mrs_proc.py”, line 746, in unlike
good, bad = preproc.shift_to_reference(dataobj.data,
TypeError: shift_to_reference() got an unexpected keyword argument ‘ppmlim’
Looking at the FSL-MRS documentation, it should be optional to provide a ppmlim for fsl_mrs_proc unlike, so I’m not sure how this error arises and how to work around it. I have also tried to provide a ppm window, but this also yields errors. Does anyone have experience with this error and/or ways to solve it?
Thanks in advance!
Best,
Daphne