Hello
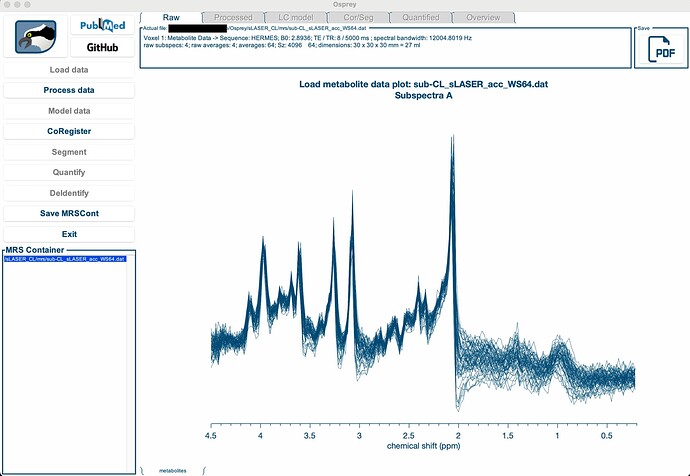
I am attempting to implement a sLASER sequence (using the Deelchand et al., 2022 ‘Plug-and-play’ method) for an upcoming pilot study. I have collected a scan but when I load it into Osprey, it thinks it is a HERMES sequence. I am able to load and see the spectra, but when I click ‘Process data’, I get an error (see screen shots below).
For reference, this is a Siemens Prisma, TE 1 = 8, TE 2 = 12, TE 3 =10, TR 5000ms, 64 averages, in the midline ACC, human not phantom, Osprey version 2.4.0.
Could this be an issue with how I set up the sequence?
Thanks!
Bethany
Hi Bethany,
Absolutely not your fault. This will be an easy fix. Osprey makes some (more or less) educated guesses from a sequence identifier string in the Twix file header. There’s a whole decision tree of if statements, and under certain circumstances it arrives at the conclusion ‘HERMES’ - I’ve seen this happen before if it doesn’t know the string.
We can probably fix this without you sharing the data:
Can you:
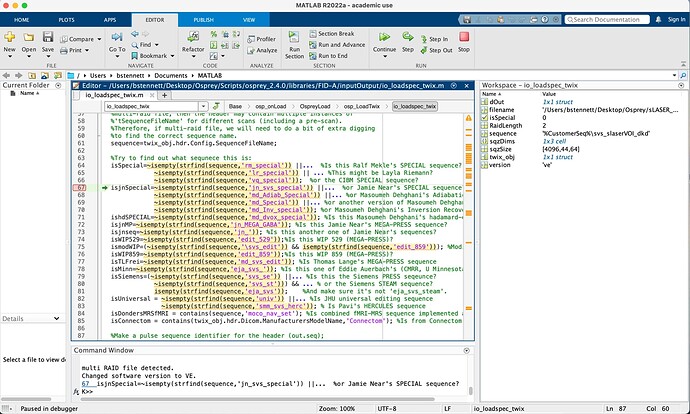
- open io_loadspec_twix in the editor
- set a sequence break point in line 67 (it should start with
isSpecial=~) - to do that, click on the line number 67 in the left gutter so that a red dot appears - execute your analysis again
- when the execution stops at this line, get the value of the variable
sequencethat is defined in line 64 - write that value here

Thanks,
Georg
Come to think of it, did the Minnesota folks share a basis set for this sequence with you?
Hi Georg,
Thank you so much for you quick reply. I am not very experienced with Matlab code, but I believe the value I received is ‘0’. I am attaching a screenshot of the editor and workspace.
I did not get a basis set from Minnesota, but I am sure that I could. I did download one from MRScloud for Siemens, sLASER, TE 8. Do you recommend I get a basis set directly from Minnesota?
Thank you!
Bethany
Hi Bethany,
I just posted a fix to the Osprey develop repository. Can you download that version (i.e. not the latest release, but the develop branch) and give it another shot?
I would recommend getting a Minnesota basis set. The MRSCloud sLASER simulation is, I believe, currently rather generic, i.e. doesn’t have the CMRR timings. I’m working on restructuring the MRSCloud code so that we can offer CMRR simulations, but for now, better to get it from them directly, especially since this is appears to be a brand new sequence.
Thanks,
Georg
Hi @admin,
I couldn’t figure out how to download from the develop brach without downloading all of Osprey again so I manually made the changes listed. Osprey successfully loaded and processed the data!
I did not make it to the model data step because I do not have a basis set to use yet. Even thought the MRScloud basis set is generic as of right now, when the time comes, would I download a TE 8 or TE 30 (8+12+10) for this sequence.
As a side note, I do have .rda and .ima files from this scan. Is there a recommended file type to use (i.e., .dat, .rda, .ima)?
Thanks again!
Bethany
sub-CL_sLASER_acc_WS64_Voxel_1_OspreyProcess_metab_A.pdf (207.1 KB)
Anything that lets you export single-transient data.
IMA should be fine for that sequence I think, Osprey can read a stack of them in if you give a folder containing them as the input argument.
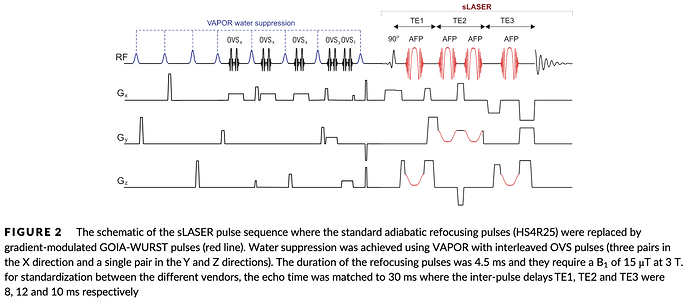
TE will always be 30 ms. The 8 ms here is a sub-echo time, so only describes the time between excitation and the occurrence of the first of three echoes (only the last one is being read out):
(https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/epdf/10.1002/nbm.4218)
Hi both,
I’m also considering using the Oz and Deechland sLASER sequence as the standard unedited technique on our Prisma. I’d be very happy to see Osprey basis sets (or scripts to generate them) available for this sequence ![]()
Cheers,
Michael
…hmm, thanks to this thread I’ve just noticed the 2022 update to this sequence!
Meanwhile, I’ve been trying to processing data from the 2016 version.
Using a similar modification to @admin Georg’s above, I can load and process the data:
… || ~isempty(strfind(sequence,‘svs_slaser_dkd’))
I get a decent fit using the Universal_Siemens basis generated from mricloud. However, water fitting doesn’t work because the basis doesn’t include H2O.
As an aside, it seems Osprey doesn’t distinguish metabolite and integrated water reference data within a Siemens VE11C PRESS twix file - I can acquire the ref data separately, of course.
Just mentioning in case it’s useful.
Thanks for providing Osprey, by the way. Really like it…
Hi @mjt320,
Thanks, that’s really kind feedback. We’ll make sure to include the svs_slaser_dkd tag in the next release, too. I’ve pushed it to the develop branch for now.
I’ve also forwarded your feedback to the folks in charge of the MRSCloud interface - H2O should really be automatically added to whatever list of metabolites the user input is.
Re: integrated water reference data: Yeah, we have not encountered data like that in our daily work yet. I don’t think it’s hard to implement (GE has integrated water reference, too).
Cheers, and thanks,
Georg
@mjt320 MRSCloud now has H2O in the list of metabolites, too - not sure whether it’s checked by default so worth including each time you submit a job!
Fantastic, thank you @admin
Michael
I have installed Dinesh semi-laser sequence with 40 averages but starting with 2 water peaks and finishing with 2 water peaks for a total of 44 acquisitions. Could you help me with the control file and basic setup ?
Sincerely
Gregory
This sequence will need to be accommodated with adjustments to the TWIX loader (io_loadspec_twix). If you can make these modifications yourself, that would be awesome (and we’d welcome a pull request). If not, please share a de-identified (or even better yet, phantom) dataset with us.