Hi there,
I am trying to analyze some 3T GE MEGA-PRES data with Osprey
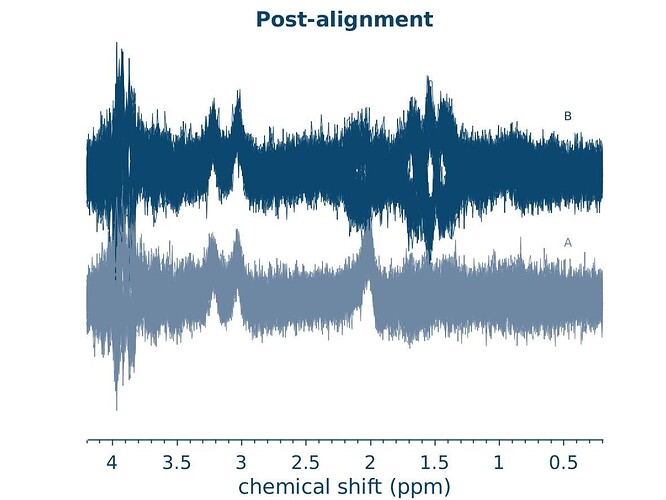
In the figures that Osprey produces, I noticed that it seems to be loading some of the spectra inverted? I hope this isn’t a very naive mistake of mine
Photo for clarity:
This is the .json I used:
{
“seqType”: “MEGA”,
“editTarget”: [
“GABA”
],
“dataScenario”: “invivo”,
“MM3coModel”: “3to2MM”,
“FWHMMM3co”: “”,
“SpecReg”: “RobSpecReg”,
“SubSpecAlignment”: “L2Norm”,
“UnstableWater”: “0”,
“saveLCM”: “1”,
“savejMRUI”: “0”,
“saveVendor”: “0”,
“saveNII”: “0”,
“savePDF”: “0”,
“method”: “Osprey”,
“ECCmetab”: “1”,
“ECCmm”: “1”,
“includeMetabs”: [
“default”
],
“style”: “Separate”,
“lolim_range”: “0.5”,
“uplim_range”: “4.0”,
“lolim_rangew”: “2.0”,
“uplim_rangew”: “7.4”,
“bLineKnotSpace”: “0.4”,
“fitMM”: “1”,
“basisSet”: “BasisSets/3TGE_MEGAPRESS_68ms”,
“files”: [
“P22016.7”
],
“files_nii”: [
“anat/sub-Rett01_T1w.nii”
],
“file_stat”: [
“”
],
“outputFolder”: [
“osprey”
]
}
Is there a fix for this?