Hi Georg,
This is exceptional, thank you so much for the detailed suggestions I am beyond grateful! Its been quite the journey getting this project off the ground.
To respond to some of your points:
- For my shimming procedure I do the auto GRE shim 2x with a shim box larger than my volume (but not close to the edges of the brain etc.). I apply the autoshim, decrease the shim box size to match that of my VOI and then do an intershim where I manually change the first order shims to see if I can knock down the FWHM a few Hz. This has gotten me the best results thus far on my machine. I will look into the CMRR spectro package.
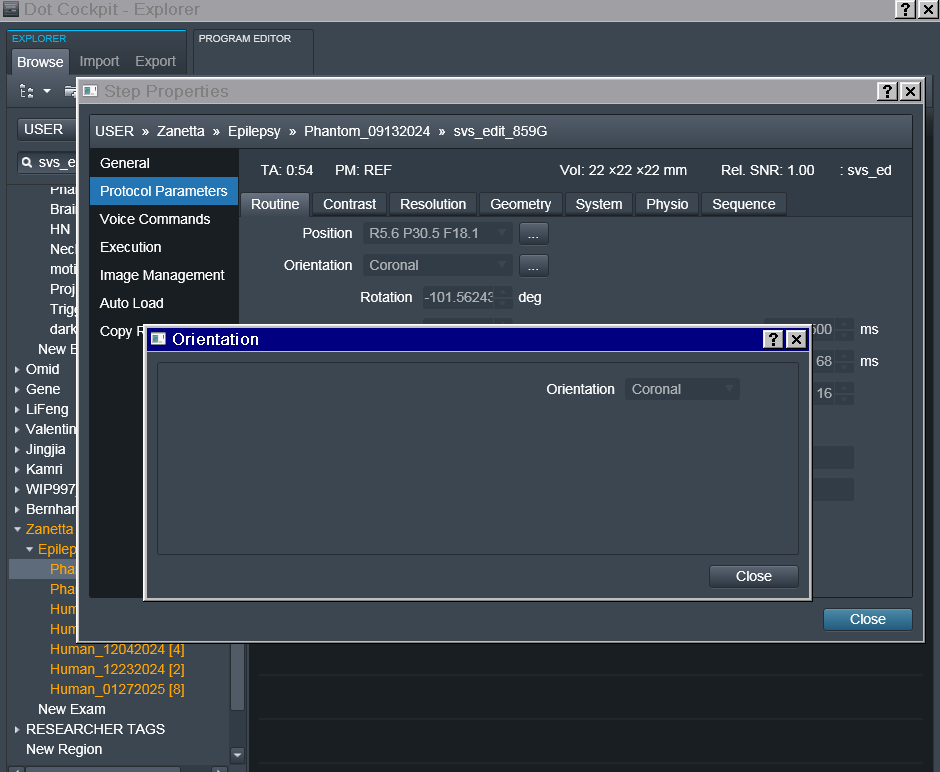
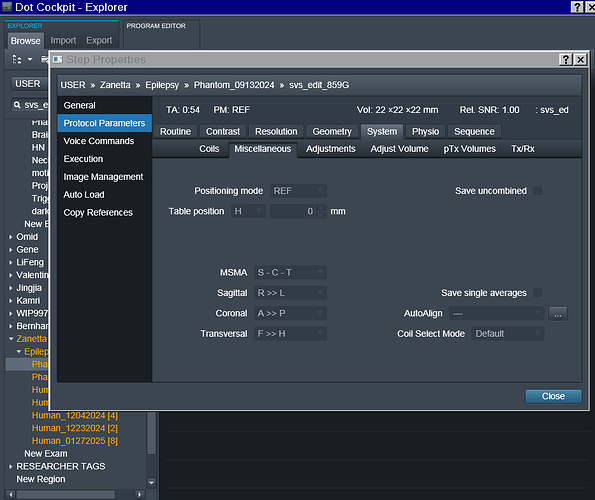
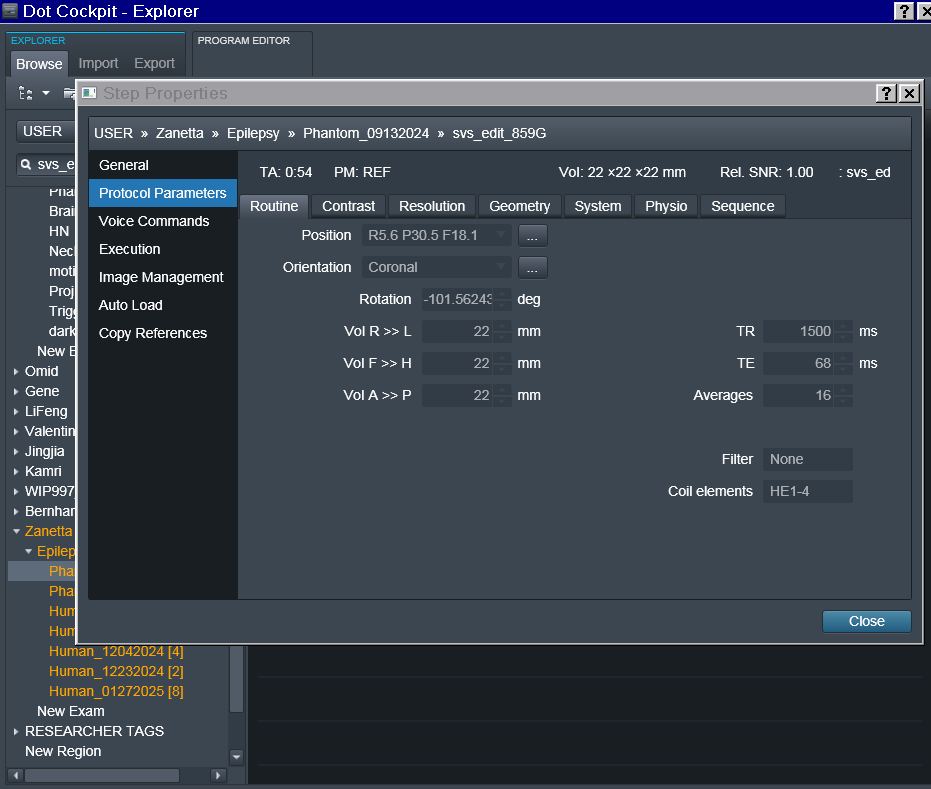
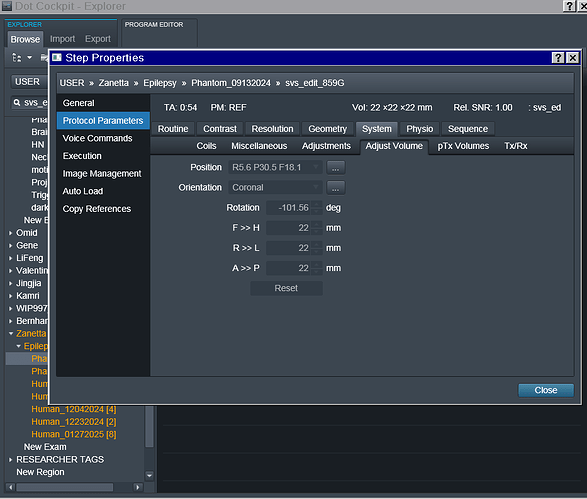
- In terms of the orientation, it is set to Coronal and greyed out so I can’t seem to change it. Screenshots of the exam card are attached.
- Ok it seems that sticking to a TR 2000 is the best option with voxel size 27 mm iso.
- I did in fact miss those lines of code. I added them and repeated the analysis. Results are here. However, should the resulting plots be labeled with “Gaba+” and not just “Gaba”? And my concern is that it quantifies a Gaba concentration of 0, even in the two svs protocols in the PCC. Attached is an example job file.
jobTwix_invivo_megapress_859G_mPFC_MMRob_EXAMPLE.m (19.1 KB) - Noted on the # of averages for the water ref scan as well as the delta frequency under Sequence → Common, it seems I missed it for the KC edit.
- So you suggest turning RFA off for all sequences? So far I have only turned it off for the water ref scan to achieve “RF OFF” water suppression. Otherwise when RFA is turned on, the only options for water suppression are “water sat.” or “weak water suppression.” This is a mostly clinical scanner so I am unsure if there are heavy duty scans run frequently on the machine. I will double check - thank you for the tip.
Thank you again for your prompt and detailed reply!
All the best,
Zanetta