Dear Experts,
I have been piloting the Siemens (Scanner: Prisma; Software version: VE11C) MEGAPRESS (859G WIP) for a few months now using Osprey for processing. Recently, I decided to run the sequence in the PCC to check that all my sequence parameters are correct and that I get the desired result. I believe acquisition in the PCC looks as expected, but its the acquisition in the mPFC or any frontal region (including ACC) that’s giving me an issue.
For my most recent data I also tried the three different fitting algorithms (Default:RobSpecReg RestrSpecReg and, ProbSpecReg) and I believe the default fits best.
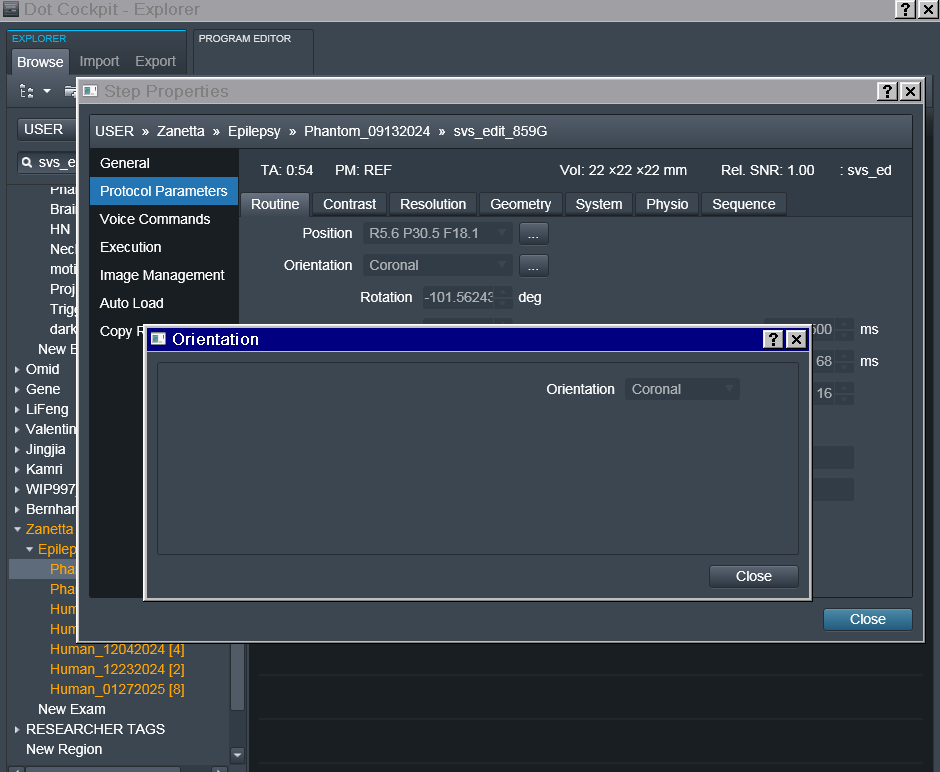
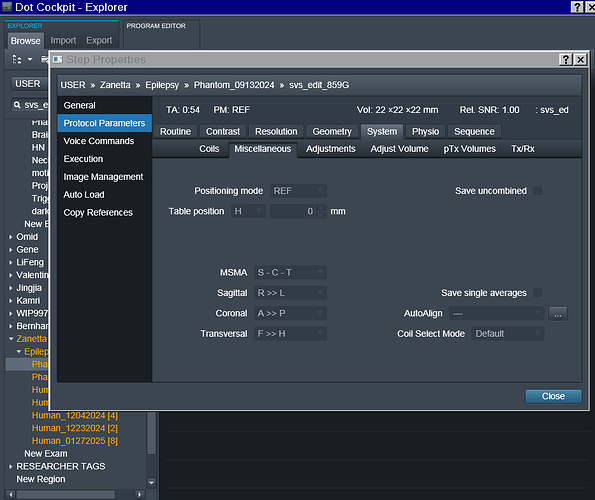
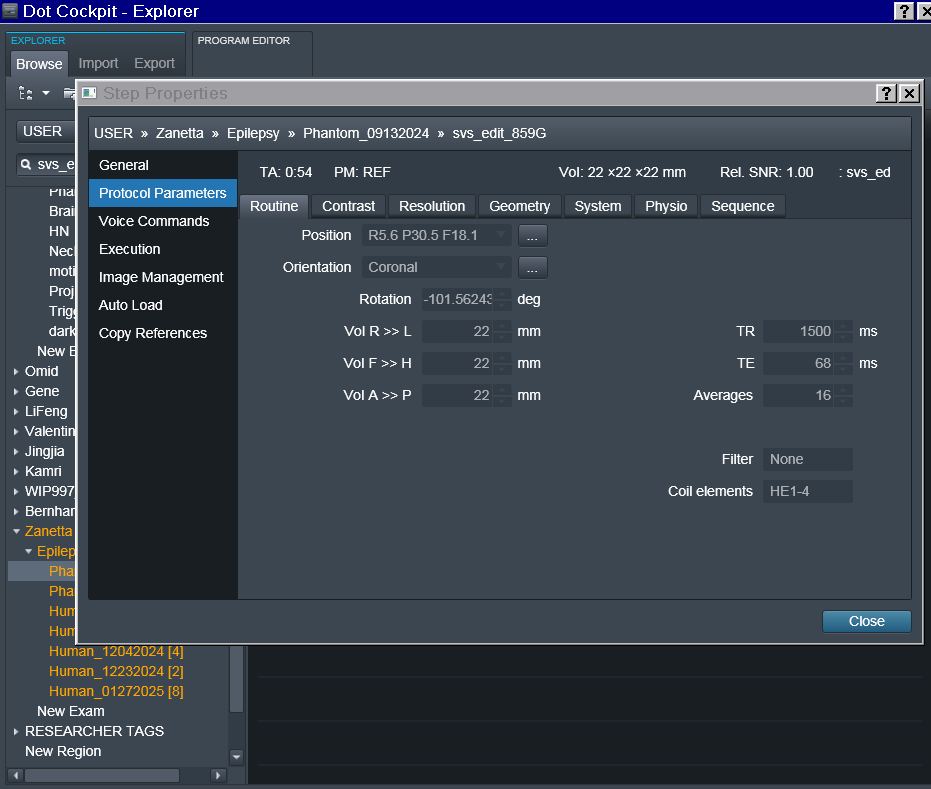
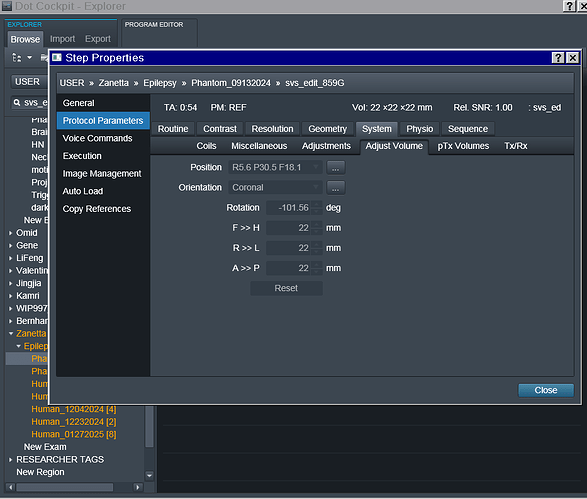
In the power point linked here I have included details regarding each acquisition, voxel localization, chemical shift PDFs (only for the RobSpecReg as the chemical shift PDFs for the other fittings were very similar), and the metab fit plots for each fit. The metab fit plots are generated using eddy correction with the water reference scan. I have also attached the entire acquisition protocol. (note: the last two acquisitions are both localized to the mPFC, I just mistakenly left the “PCC” naming during the scan.)
The KC Edit acquisition was one used by other researchers in my department, so I just wanted to test it out to see if I get good results.
For SVS Edit 1 & 2, I went off of another post on MRS hub to optimize the parameters. I think these two acquisitions are quite comparable. Ideally I would use a shorter TR and # of averages in my final acquisition as I need to decrease my scan time as much as possible.
SVS Edit 3 is exactly like SVS Edit 2 but just localized in the mPFC. This is where I struggle the most and don’t know what to do in order to improve the fit. The chemical shift is of poor quality, the data is quite noisy.
I know there are a few other things that could be contributing to this noisy data (shim, motion). I did an interactive shim and got it down to 20.4 Hz in the mPFC. Admittedly, the other shims were below 14 Hz for the PCC/OCC, but it was simply not possible to go down to that value in the mPFC. This was also the last picture that I took, so perhaps there is some motion artifact here but I am not sure. Right before this image I acquired two PRESS sequences also in the mPFC and the data came out very clean.
I am unsure how to proceed. Ultimately, I need to localize in the frontal region and I wonder what else I can do to improve the data? Any and all suggestions would be highly appreciated. THANK YOU!
Regards,
Zanetta
Human_01272025.pdf (114.1 KB)