Thanks for a very nice program, I really appreciate the effort to put all pieces together to form a complete pipeline for MRS.
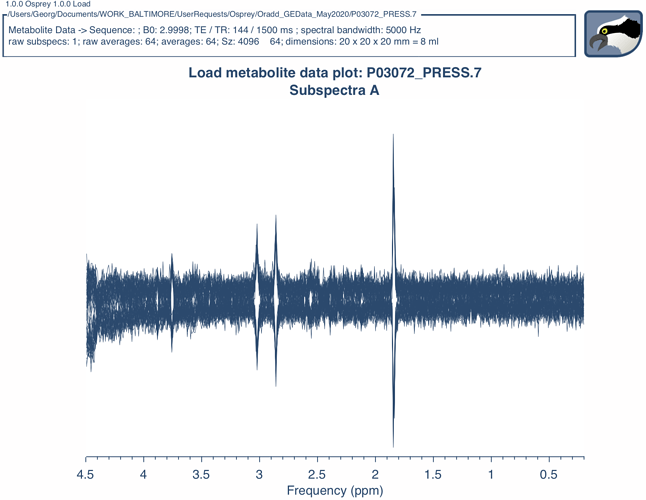
I have recently installed the GE WIP MRS package, including among other things GABA editing, on our GE MR750 scanner and wanted to try Osprey on the MRS phantom provided by GE (GE braino, containing NAA, Cho, Lac, Glu, Cre and myoinositol). My problem is that the metabolite basis set provided with the Osprey package (/ge/press35 and ge/megapress_gaba68) is made for 37 degrees, while the phantom is at room temperature (20C), so the chemical shifts are no longer valid. Do you by any chance have basis sets suitable for phantom data?
/Greger
Hi Greger,
Thank you very much for your inquiry. We have received your example phantom dataset via e-mail, and we’ll do some testing to incorporate a phantom data processing and modelling pathway into Osprey. The basis sets will remain the same, it’s just a matter of referencing and phasing the data appropriately to account for the temperature-related chemical shift.
I’ll post an update to this topic once we have added the feature.
Best wishes,
Georg
Hi Greger,
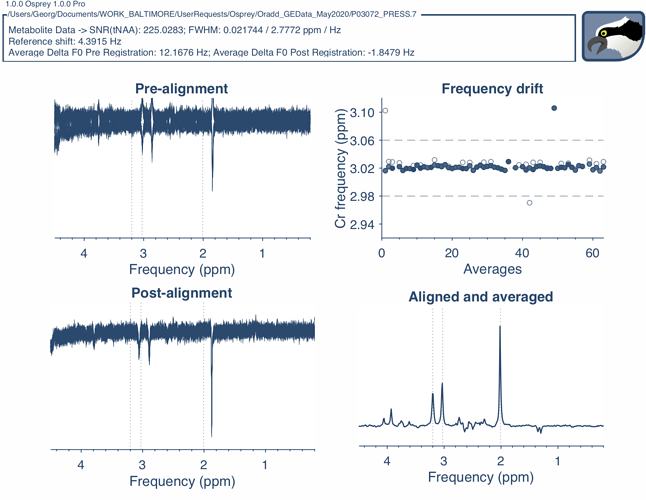
We’ve made some progress on loading and fitting phantom data, with a couple adjustments that I had to branch out in the workflow. For starters, your data seemed to have an exquisitely great shim, and my simulated basis set couldn’t keep up with it! I threw some linebroadening on the phantom data, and voila, it worked well. I’ve also introduced the option to define the metabolites you want to include in your basis set in the job file.
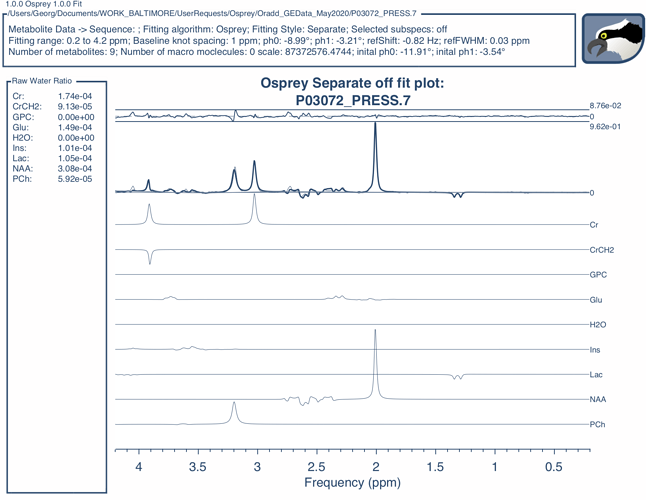
I don’t know what kind of choline is in the braino phantom - the fit there does look a little off, and neither GPC nor PCh might be perfectly suitable to analyze phantom data.
Next stop will be the MEGA-edited data you sent. Let’s see whether this works as smoothly. I’ll let you know in this thread as soon as I merge these new developments into the Osprey GitHub repository.
Cheers, best wishes, and thank you for your interest in using Osprey,
Georg
Hi Georg,
Thanks for your efforts, the fitting looks good, and I hope that the edited data will also come out smoothly.
The choline in the braino phantom is “naked” choline, (CH3)3-N-CH2-CH2-OH in the form of cholinechloride, so it is actually a good measure of the fitting routine that it got problems with fitting it to GPC and/or PCh. I could probably find the correct spin system params and simulate the lineshape once I have figured out how to do the simulations.
Just out of interest, what did you do to adapt to the phantom data? Did you just change the resonance frequency of water or did you adapt the params for all involved spin systems?
Cheers,
Greger
Ah, yes - it might be worthwhile for us to dig out the free choline spin system and add a FID-A definition. The unmodeled 4.05 ppm peak in the figure above also checks out to be free choline. Let me know if this is important for you, and I’ll re-run the simulations.
Honestly, most of it came together pretty easily once I shifted the spectrum by 0.15 ppm, stiffened the baseline (knot spacing of 1 ppm), and threw the linebroadening on top. These settings are all behind a ‘phantom’ switch now, so in-vivo data are processed as usual.
Cheers,
Georg