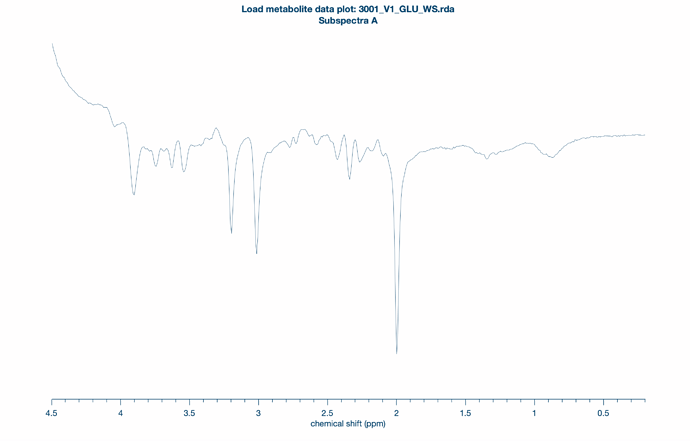
Hello,
First of all, thank you for providing this software! It is very well done and easy to use. I looked through the other posts and didn’t see anything matching my concerns, but if for some reason I overlooked a conversation please feel free to point me in that direction.
I began working through the pipeline using 1 participant (water suppressed and non-water suppressed .rda files and T1.nii file) and ran into some things/warnings that made me want to get an expert opinion on. I am able to run through the full pipeline, but I wanted to make sure that nothing described below would be messing up the final analysis. I am using Monterey 12.1 and Matlab R2020b.
-
When I load the data, the preprocessed raw spectrum is inverted. This is corrected after going through the processing step.
-
After processing, there is no data for the chemical shift drift.
-
I am getting warnings after both the model and quantify steps. I had to upload them as txt files because it wasn’t letting me post as it thought I was tagging users since the @ is used in both warnings.
Model Warning
Model Data_Warning.txt (3.7 KB)
Quantify Warning
Quantify_Warning .txt (4.9 KB)
Thanks is advance!