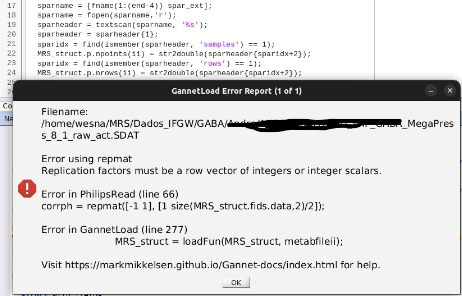
This is one of the .SPAR data. Is this enough to double-check?
My idea would be to try changing the value of the ‘rows’ field to 104 and then changing the .SDAT as well.
{'examination_name': 'fMRS-GABA-3freqs',
'scan_id': 'GABA_MegaPress',
'scan_date': '2012.03.19 16:42:11',
'patient_name': '',
'patient_birth_date': '',
'patient_position': 'head_first',
'patient_orientation': 'supine',
'samples': 2048,
'rows': 105,
'synthesizer_frequency': 127770872,
'offset_frequency': 0,
'sample_frequency': 2000,
'echo_nr': 1,
'mix_number': 1,
'nucleus': '1H',
't0_mu1_direction': 0,
'echo_time': 68,
'repetition_time': 2000,
'averages': 8,
'volume_selection_enable': 'yes',
'volumes': 1,
'ap_size': 30,
'lr_size': 30,
'cc_size': 30,
'ap_off_center': 60.34542465,
'lr_off_center': -5.00035429,
'cc_off_center': -5.641904354,
'ap_angulation': 1.679412961,
'lr_angulation': -38.76796722,
'cc_angulation': 0.7264128327,
'volume_selection_method': 1,
'phase_encoding_enable': 'no',
't1_measurement_enable': 'no',
't2_measurement_enable': 'no',
'time_series_enable': 'no',
'image_plane_slice_thickness': 0,
'slice_distance': 0,
'nr_of_slices_for_multislice': 1,
'Spec.image in plane transf': 'plusA-plusB',
'spec_data_type': 'cf',
'spec_sample_extension': '[V]',
'spec_num_col': 2048,
'spec_col_lower_val': -1000,
'spec_col_upper_val': -998.9760132,
'spec_col_extension': '[sec]',
'spec_num_row': 105,
'spec_row_lower_val': 1,
'spec_row_upper_val': 105,
'spec_row_extension': '[index]',
'num_dimensions': 2,
'dim1_ext': '[sec]',
'dim1_pnts': 2048,
'dim1_low_val': -1000,
'dim1_step': 0.0005000000237,
'dim1_direction': 'mu1',
'dim1_t0_point': 0,
'dim2_ext': '[num]',
'dim2_pnts': 1,
'dim2_low_val': 1.0,
'dim2_step': 1.0,
'dim2_direction': 'x',
'dim2_t0_point': 50,
'dim3_ext': '[num]',
'dim3_pnts': 1,
'dim3_low_val': 1.0,
'dim3_step': 1.0,
'dim3_direction': 'y',
'dim3_t0_point': 50,
'echo_acquisition': 'ECHO',
'TSI_factor': 0,
'spectrum_echo_time': 68,
'spectrum_inversion_time': 0,
'image_chemical_shift': 0,
'resp_motion_comp_technique': 'NONE',
'de_coupling': 'NO',
'equipment_sw_verions': '2.6.3 ; .6.3 ;',
'placeholder1': None,
'placeholder2': None}
![]()