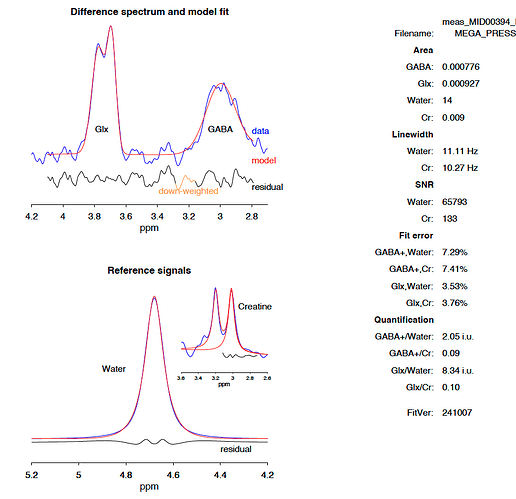
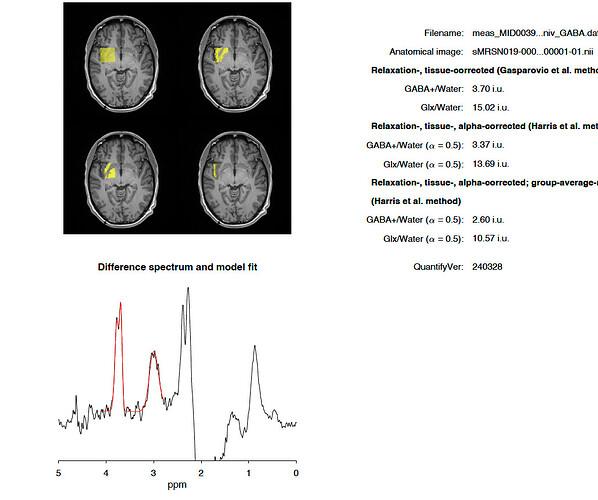
I have run a batch of 88 scans in Gannet and excluded poor quality scans. However, I have noticed a few scans (one in particular) which have a nice spectrum, sensible GABA and Glx results, but a high Glu result (far higher than Glx). Do you know what the cause might be?
As an example, the results from this scan attached for GABA and Glx are sensibly distributed in my data, but the output.csv file gives a Glu conc. of 147.505, 167.43, 265.592, 241.942, and 186.849 respectively for i.u., CSF-corr, +tissue-corr, +alpha-corr and +group-norm respectively. The median concentration is ~8 i.u.
Glu_FWHM 3.297, Glu_SNR 207.246, Glu_fit_error 0.101, Glu_fit_error_Cr 1.429, and Glu_fit_error_w 0.593.
I know that reporting Glu separately has its limitations, but I am surprised that the figures are far higher than the Glx. I can report Glx only, but I wanted to check I wasn’t missing anything.
Many thanks!