I fail to get a good fit and would greatly appreciate any help with basis spectra and sequence description.
Thanks in advance
Hi @Sipan , is this fsl-mrs? If so, could you upload the html report output? Guessing what is going on from just this output is hard.
@wclarke
Thank you for your feedback, yes its done by fsl-mrs. I was able to follow the practical and get the exact fit for the practical data, but when I tried my own data it did not fit well. Please find my report (pdf) attached.
report.html.pdf (1.1 MB)
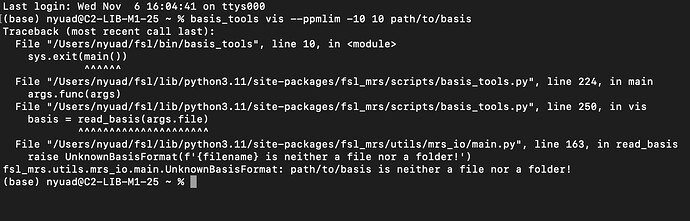
What basis are you using? Can you use basis_tools vis --ppmlim -10 10 path/to/basis and send the output?
Maybe try updating /path/to/basis to the path to your basis directory?
You mean the program is using the practical’s basis?
I wanted to ask another question, I if I use 3T scanner and PRESS sequence, can I just change coherence filter and filed strength in the practical’s sequence description and use it, or I need to write my own sequence description from 0.
Am I right in thinking you are using the basis fromt he practical to try and fit your data? And that your data is from a 3T, PRESS sequence? Is the sequence a vendor one or from another source? E.g. from the CMRR spectro package.
You would need to make modifications beyond the coherence filter and field strength, but possibly not much more. Depends on your answers above.
Yes you are right
I use the basis and sequence description from practical
The data is from 3T Siemens MAGNETOM Prisma MRI scanner
The sequence is PRESS I think from the vendor
I greatly appreciate your help!
Dear @wclarke I would more than greatly appreciate your feedback
Thanks in advance
What echo time do you use? Does it match any that are listed here? https://juchem.bme.columbia.edu/content/mr-spectroscopy-basis-sets
If so , I would recommend just using one of these.
Echo time is 30ms, which is available. Thank you so much for your help!
Dear @wclarke is there a command/tool to combine and convert those .raw files into Json file (basis spectra) to be readable by FSL. The Columbia’s website says that the files are readable by LCModel and there was no mention of FSL and also I did not found such command in the FSL practical. If you could help with modifying the description provided by practical for my data I would be grateful!
Thanks in advance
Hi @Sipan ,
You can use the following command:
basis_tools convert path/to/my/lcmbasis.BASIS path/to/my/fslbasis
Convert LCModel (.Basis), LCModel (directory of .raw) or JMRUI format basis sets to FSL-MRS (.json) format.
Note that the bandwidth and fieldstrength must be supplied manually to the CLI for the .raw format.
This is from the instructions here Basis Spectra Manipulation — FSL-MRS 2.3.2 documentation
Sorry on my mobile today so can’t do as many details. Take a look at the cli help.
Thanks a lot, I will try
Dear @wclarke
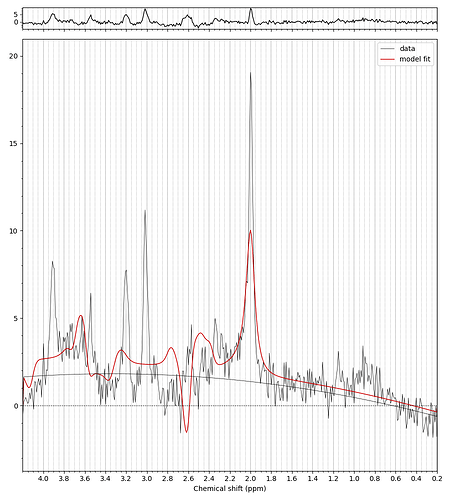
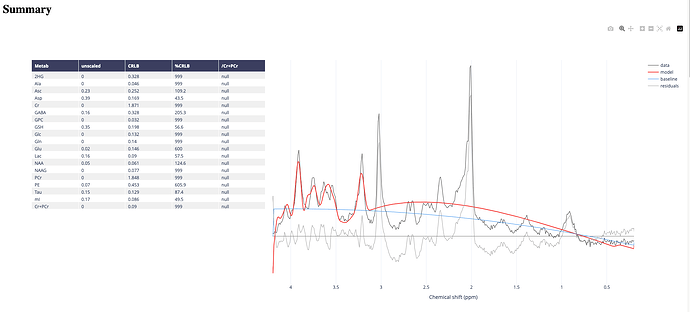
I used the data set you provided to generate my basis, which was successful for metabolites, however, the dataset misses the set for macromolecules, which I think is really important, as it appears along all spectrum. Can you please validate my reasoning that the failure to get a nice peak occurs due to absence of macromolecules data set in my basis. Please see the image of basis and fitting and the pdf of report attached.
I would greatly appreciate your feedback
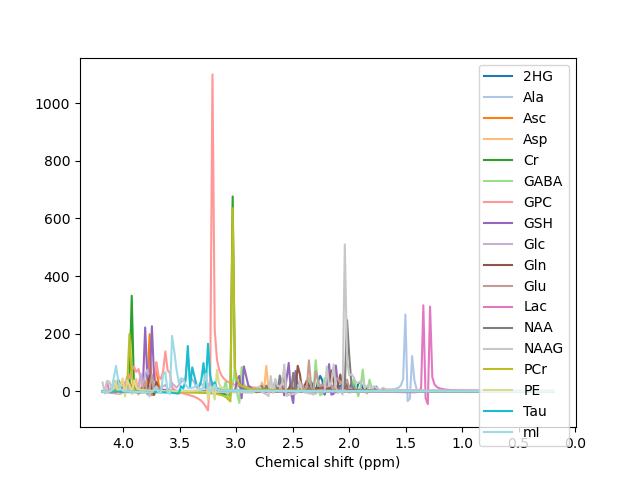
report.pdf (146.1 KB)