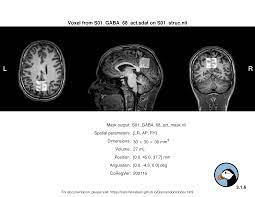
Hi Mark,
Thank you for the reply. Currently I am trying to use CoRegStandAlone.m. However I am having difficulty loading in my SPAR/SDAT data. The codes that I tried are posted below.
Example1:
data_dir = ‘/Data PVC/Gannet/Gannet-main/Gannet-main/testdata/test’;
metab = {‘test_SVS_14_2_raw_act.sdat’};
metab = fullfile(data_dir, metab);
water = {‘test_SVS_14_2_raw_ref.sdat’};
water = fullfile(data_dir, water);
anat = {‘test_T1w.nii’};
anat = fullfile(data_dir, anat);
MRS = CoRegStandAlone({metab,water},{anat,anat});
Example 2:
MRS = CoRegStandAlone({‘test_SVS_4_2_raw_act.SDAT’,‘test_SVS_4_2_raw_ref.SDAT’},{‘test_T1w.nii’,‘test_T1w.nii’});
Both of these codes do not seem to work. As I do not understand how I can use both my SPAR and SDAT data, which I feel like is needed to make this work properly.
This is listed inside the function:
% MRS_struct = CoRegStandAlone(metabfile, struc)
"Input:
% metabfile - cell array containing the path(s) to an MRS data file
% in one of the following formats: Philips SDAT/SPAR,
% Siemens TWIX (.dat), Siemens RDA (.rda), GE (.7), or
% DICOM (*.ima, *.dcm).
struc - cell array containing the path(s) to either a NIfTI
% file (*.nii) (for Philips and Siemens data) or a folder
% containing DICOMs (.*dcm) (for GE data).
Kind regards,
Sander