please drop me an message with your email address.
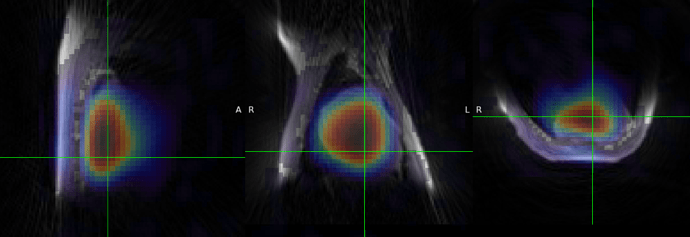
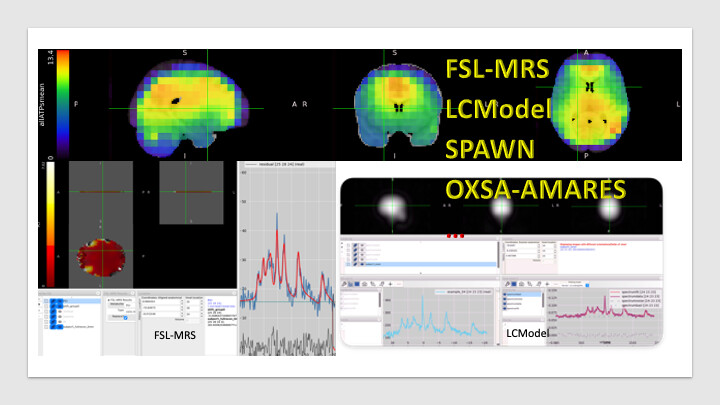
We have been working on lcmodel, fsl-mrs(@wclarke helped us to transfer lcmodel basis set to fsl-mrs) and spawn for 31P MRSI. With our lcmodel pipeline, our 3D novel rosette ute 31P MRSI apporach got 2nd place at the ismrm reproducibility challenge. We recently tested the 3D novel rosette ute 31P MRSI on the bruker system too (if you are interested in ![]() )
)
You can find more details in th following GitHub page.
lcmodel data analysis pipeline please use the following link.