I think I find a coding fault that may create error while utilising OspreyFit, and I’d like to share it.
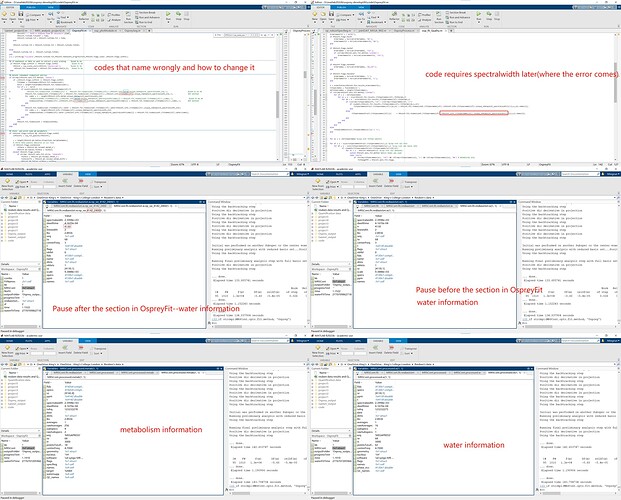
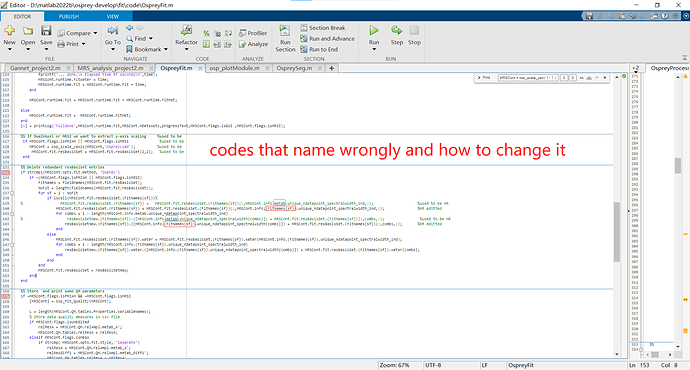
OspreyFit has a section around line 130 that starts with “%% Delete redundant resBasiset entries” and rewrites one struct called “MRSCont.fit.resBasisSet.” It accesses water and metabolite data using FitNames{sf}(sf=1, sf=2), but only acquires metabolite data via “MRSCont.info.metab.”. To be more explicit, it uses the spectralwidth of metabolite to rename both cells (“w” and “metab”) in the struct (“MRSCont.fit.resBasisSet”).
So when another function named “osp_fit_Quality” requires these two cells again using the spectralwidth from water and metabolite respectively, the intended water spectralwidth can’t match with the cell name of water information ( because it names by metabolite spectralwidth), and it will shows the error “unrecognized field name np_sw_actual water spectralwidth” (for my data, it shows “unrecognized field name np_sw_4130_2400”, and the water is named “np_sw_4142_2400” by metabolite information)
I tried adding a pause before and after the part “%% Delete redundant resBasiset entries” to analyse my data, which clearly demonstrates that the water is named by the spectralwidth of metabolism (it is “np_sw_4142_2400”, but should be “np_sw_4130_2400”)
Although there could be a more elegant solution to this problem. By changing MRSCont.info.metab to MRSCont.info.(FitNames{sf}) according to the first screenshot i shared, the problem can be fixed quickly.
I have checked that the recent OspreyFit code they update recently and they haven’t make change to these codes. If anyone meet the similar situation, you can save your time by editting these codes.