@Helge @admin
I took a look at the SummaryMRSinMRS.md file that was created.
You may need to add information that was not avaialble in the raw data.
|1. Hardware| |
|--|--|
|a. Field strength [T]| 3 T|
|b. Manufacturer| GE|
|c. Model (software version if available)| Rev_number �|
|d. RF coils: nuclei (transmit/receive), number of channels, type, body part| 1H |
|e. Additional hardware| -|
|2. Acquisition| |
|--|--|
|a. Pulse sequence | press|
|b. Volume of interest (VOI) locations | -|
|c. Nominal VOI size [mm<sup>3</sup>]| 28 x 28 x 28 mm<sup>3</sup>|
|d. Repetition time (TR), echo time (TE) [ms]| TR 2000 ms, TE 35 ms|
|e. Total number of averages per spectrum <br> i. Number of averaged specra per subspectrum | 4 total averages with 4 averages per subspectrum|
|f. Additional sequence parameters | F1: 5000 Hz, 4096 points|
|g. Water suppression method | -|
|h. Shimming method, reference peak, and threshold of acceptance of shim chosen | -|
|i. Trigger or motion correction| -|
|3. Data analysis methods and outputs| |
|--|--|
|a. Analysis software | Osprey 2.4.0|
|b. Processing steps deviating from Osprey | None|
|c. Output measure | tCr, rawWaterScaled, CSFWaterScaled, TissCorrWaterScaled
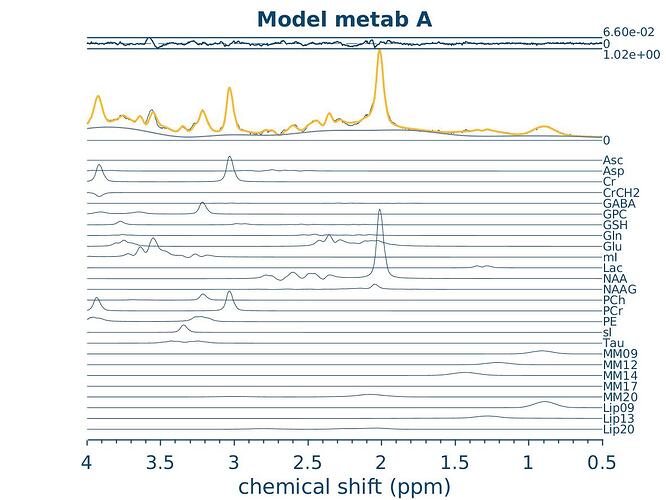
|d. Quantification references and assumptions, fitting model assumptions| Basis set list:<br> AscAsp,Cr,CrCH2,GABA,GPC,GSH,Gln,Glu,mI,<br>Lac,NAA,NAAG,PCh,PCr,PE,sI,Tau,MM09,<br>MM12,MM14,MM17,MM20,Lip09,Lip13,Lip20,tNAA,Glx,<br>tCho,tCr <br>Fitting method: Osprey basline knot spacing 0.40 ppm
|4. Data quality| |
|--|--|
|a. SNR (Cr), linewidth (Cr) [Hz] | SNR: 125 +- 0, linewidth 7.36 +- 0.00 Hz|
|b. Data exclusion criteria | None|
|c. Quality measures of postporcessing model fitting (Mean Relative Amplitude Residual) | 9.32 %
|d. Mean spectrum created with OspreyOverview| Figure 1
It seems to think the sequence is PRESS?
But the basis set says it’s semi Laser… weird…
Could this be the culprit?
![]()