Hi,
I am using Osprey for analyzing my SVS data acquired by STEAM and sLASER at 3T and 7T. I am facing a problem and I do not know why this is happening. I would appreciate it if you could help me.
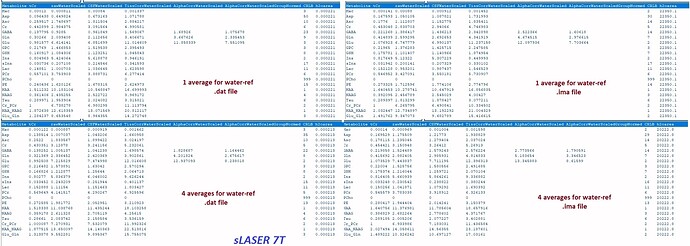
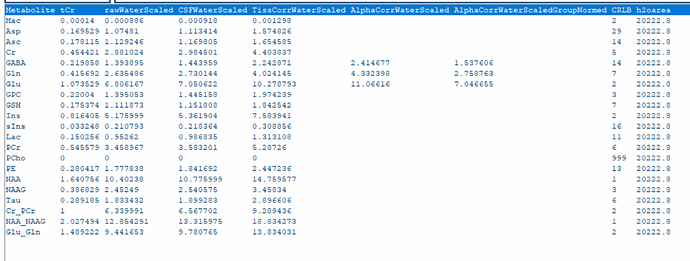
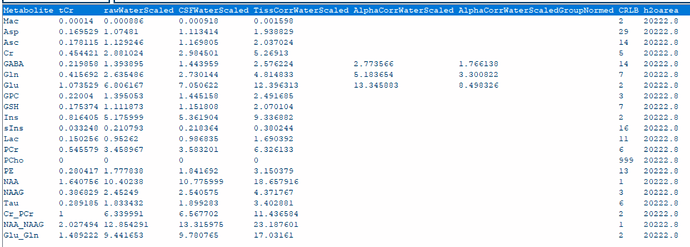
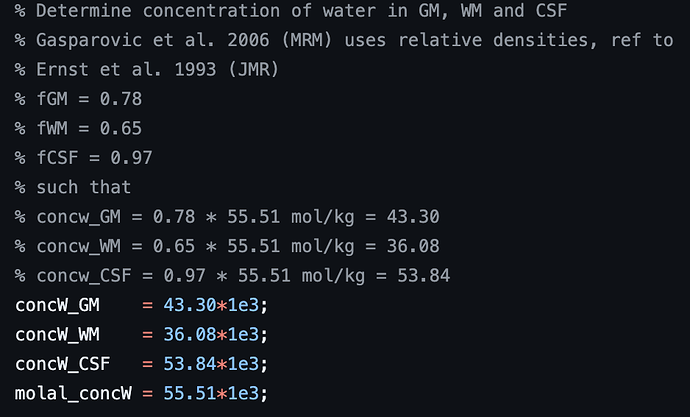
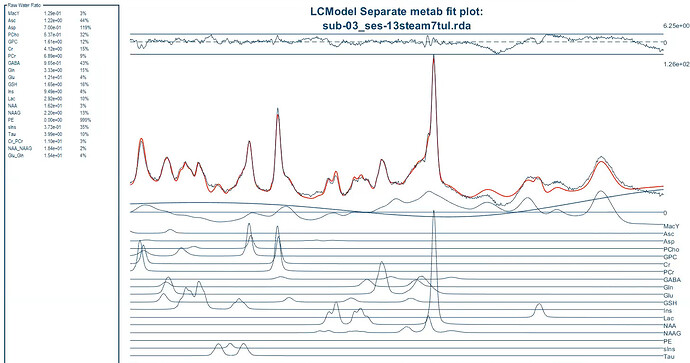
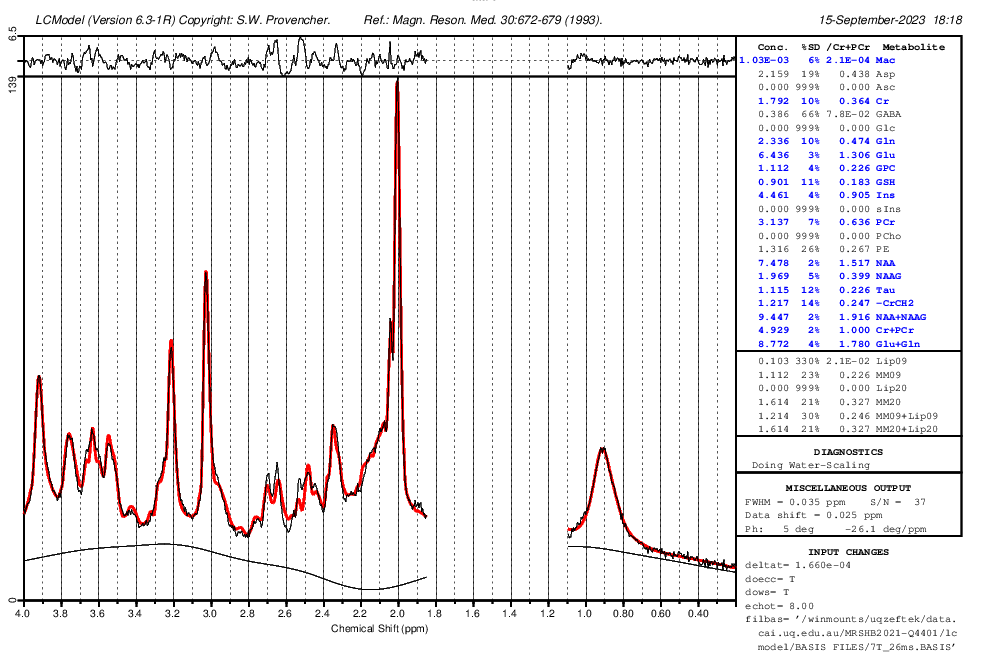
1- All TissCorrected values for both steam and slaser at 3t and 7t are higher than the normal range when I use .ima and .rda formats. I tried with dat format just for steam at 7t, and the values were in the correct range. However, my problem is I do not have the same type of format for all my data.
Do you know what should I do so that the values be in the correct range for the .ima and .rda formats? (I put screenshots of quantification tables and a couple of videos to see the quality of fitting and the procedure).
https://filesender.aarnet.edu.au/?s=download&token=41c8b385-333a-43bb-8f13-f373eb693845
2- For STEAM at 3T and 7T, I have dat and rda.
For sLASER at 3T and 7T, I have .ima and rda. All 4 Water reference scans and 32 averages for metabolite data were saved in one dat file for each sLASER, and I do not know how can I separate them. Also, I want to know if it is possible to extract the first 16 averages out of 32 from a dat file and save it as a new file?