hello,
every time that we try to acquire hercules (2019 version) data in V1, we see huge lipid/macromolecule contamination – are there any tricks to placing a 3 x 3 x 3 voxel in V1 that avoid contamination?
Hi @sdw,
Can you share screenshots of your voxel placement? What scanner are you using (GE, Philips, Siemens…)?
Depending on your voxel placement, you can either move it further away from the meninges or flip its orientation to change the order of the slice-selection pulses (as shown here) — to do this differs by the scanner manufacturer.
But more info on your data is needed to determine what may be the issue.
Mark
thanks for your help! We are using a Siemens 3t scanner.
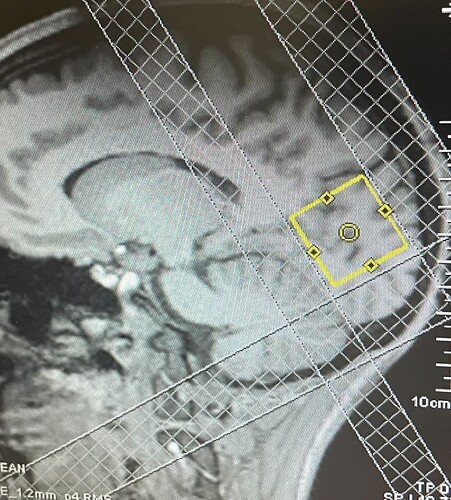
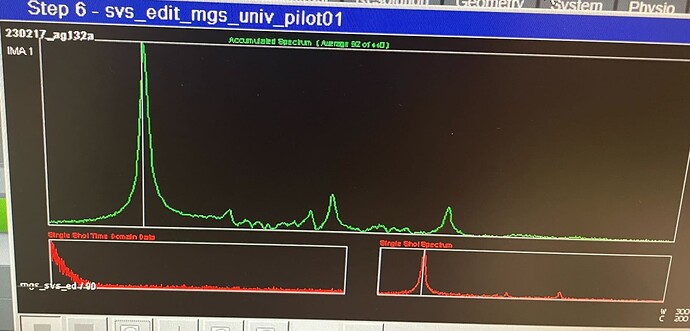
Here are 2 voxel placements - one super close to the meninges and one placed farther away that still shows something weird happening ~ 1-1.5 ppm
svs_Voxel_1_OspreyFit_sum_example2.pdf (196.1 KB)
svs_OspreyCoregSeg_example1.pdf (340.1 KB)
svs_OspreyFit_sum_example1.pdf (178.4 KB)
svs_Voxel_1_OspreyCoregSeg-example2.pdf (282.5 KB)
Standard Siemens MRS acquisitions tend to have more lipid contamination because, I believe, outer-volume suppression (OVS) is turned off by default. We saw this in our Big GABA study.
I suggest first checking whether OVS is turned on in your protocol.
thanks so much for your help! Do you happen to know where in the program card ‘OVS’ can be turned on or know if there are details posted elsewhere about how to do this? I flipped through the program cards on the scanner console and didn’t see a checkbox to turn OVS on
I’m not a Siemens user, but I think OVS is set on the “Geometry” window. I think it’s simply called “saturation” or “sat.”. But I defer to the Siemens users on this forum.
by chance, is this what you were referring to? In the geometry panel we added 3 saturation bands. the lipids we’ve been seeing did seem to disappear
Yep, that’s what I meant. I agree that the lipids have been suppressed substantially.
I would leave some space between the sat band and the voxel, otherwise you’ll risk deleting some signal inside the voxel due to the chemical shift displacement artefact (which is obviously also valid for the sat band).
What frequency is the sat band set to? IIRC, it needs to be set to -3.4 ppm (to target lipids at 1.3 ppm) since Siemens calculates it relative to water at 4.7 ppm.