I frequently review MRS data acquisition protocols for Siemens users, especially those who are new to MRS. The ‘default’ exam card settings provided by Siemens are not particularly well optimized, and I often find things that can be improved upon.
This thread is an attempt at consolidating these tips and considerations into a single ‘live’ resource to be updated over time. If you feel like something needs to be added or modified here, please post.
Basic spectral parameters and SNR considerations
-
Number of transients/averages & voxel size. The question that people ask the most about by far. Understandably - in an ideal world, we’d all scan 4 brain regions with high spatial specificity in a three-minute scan so that we can still add all the fMRI and DTI scans afterwards. My answer will always and forever be “Don’t compromise on voxel size unless you absolutely HAVE to”. This is especially true if you want to estimate low-concentration coupled signals e.g. with MEGA experiments, which we usually recommend to run in voxel sizes of 25 ml or more. If you decide to decrease your voxel size, you pay a hefty price that you cannot get a discount on by just scanning longer. Keep in mind that a voxel with 2 cm x 2 cm x 2 cm = 8 ml is less than a third of the volume of a 27-ml voxel, which means that you’d have to scan for about 10 times as long to maintain the same SNR (because SNR only increases with the square root of the number of transients, but linearly with the voxel volume). If you absolutely need to go towards lower voxel size, pilot smaller voxels in small increments (maybe 3-4 ml at a time), collect the data in a couple of volunteers, and get a sense for how much your standard deviation across the test cohort increases. We don’t always have a great sense about when your SNR will cause things to fall apart. (personal opinion piece: I will take good-SNR data that I can have faith in, even if they will require me to include a discussion paragraph about potential partial-volume effects when I write the paper, over bad-SNR data that I can’t properly analyze or interpret - any time of the day.).
-
Choose the spectral width so that the frequency range covers everything you need. At 3T, choose at least 1200 Hz (as an example, 1000 Hz at 3T means that you only get 500 Hz either side of water, i.e. slightly more than 4 ppm, but some signal-free range of the spectrum is useful). We typically choose the number of points and spectral bandwidth so that the total acquisition time is more than 500 ms (e.g., 2048 pts @ 2 kHz or 1024 pts @ 1.2 kHz), making sure that the signal has decayed completely into noise.
Acquiring multiple regions
- If you acquire MRS data from multiple regions, make sure that spectral parameters (TR, TE, spectral width, number of points, shim settings, etc.) agree between scans. It’s easiest to set up a complete and correct protocol (including water references, see below) for one region, then copy and paste the entire block to adjust voxel geometry and number of transients for the next region.
Water reference scans
It is strongly recommended to acquire water reference data for eddy-current correction and water-scaled quantification. I generally recommend to acquire one water reference with the exact same sequence used for the metabolite data (which is then used for eddy-current correction), and to acquire a separate one from the same region but with the shortest possible echo time (for the best quantification, as it has the least amount of T_2 weighting).
-
On many older software versions, you need to acquire separate water reference scans, i.e., you will copy the water-suppressed scan and change a couple of parameters.
- All separate water reference scans need to be acquired with
Sequence→Common→Delta Frequencyset to 0.0 ppm. This is to ensure that the unsuppressed water signal is co-localized with the center localization of your water-suppressed acquisition. - You will need much fewer transients (‘averages’). 4 is plenty. 2 start-up scans are plenty, too.
- For the TE-matched scan intended for eddy-current correction, set
Contrast-Common-Water SaturationtoRF off. This ensures that the water suppression gradients are still played out, which is key for accurate eddy-current correction. - For the short-TE scan intended for quantification, you can set
Contrast-Common-Water SaturationtoNone. This ensures that water suppression is de-activated entirely. - Make sure that all your scans from one single region of interest (metabolite, eddy-current reference, quantification reference) have the same number of points and spectral width and, of course, have the exact same voxel geometry.
- All separate water reference scans need to be acquired with
-
Newer product sequences (and some custom-built research sequences like from the CMRR) come with options to automatically acquire water reference scans.
- This option is activated by changing
Sequence→Common→Ref. Scan ModefromOffto one of the following options:Inline Correction(acquires ref scan, applies ECC and stores only ECC-ed data),No Inline Correction(acquires ref scan, does not apply ECC, stores ref scan and uncorrected data), orSave all(acquires ref scan, does not apply ECC, stores ref scan, uncorrected data, and corrected data). (per Siemens: Where is the non water-suppressed spectra? - Ask Anything! - MRSHub Forum)
- This option is activated by changing
Avoiding lipid contamination and out-of-voxel echoes
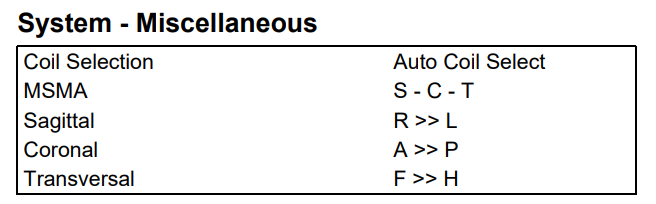
Some regions are more difficult to get good MRS data from than others. Posterior cingulate data will almost always look great. Spectra from the frontal cortex, hippocampus, or the brain stem can be more challenging - the main problems are getting a good shim, avoiding lipid contamination, and avoiding out-of-voxel echoes. Lipids and OOVs can be reduced by finding the optimal voxel geometry. Unfortunately, Siemens does not offer a visualization of a ‘chemical shift box’ like Philips does, so figuring out how to avoid lipids is less intuitive.
- Take time to pilot scans in difficult regions. Experiment with the order (MSMA) and orientations of the slice-selective gradients. Acquire all possible permutations of these parameters, each with a low number of transients. Look at the data and compare which gives low lipids and/or low out-of-voxel signals. See this thread and this paper.
Experimental order
- If you perform spectral-editing experiments such as MEGA-PRESS, it is important that your scanner does not exhibit ‘frequency drift’, i.e., a change of the main Larmor frequency over time. This can be caused by driving gradient-heavy sequences (e.g. DTI). Scanner drift varies a lot between scanners. Unless you have a very good sense how stable your scanner is, you should avoid
- running your edited MRS experiments after fMRI, DTI etc. scans
- scanning on a cold scanner first thing in the morning (some scanners have an ‘eco’ overnight mode that causes them to drift in the first 30-60 minutes or so after being used)