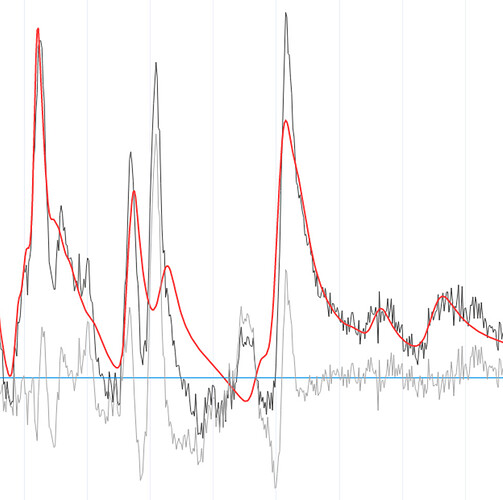
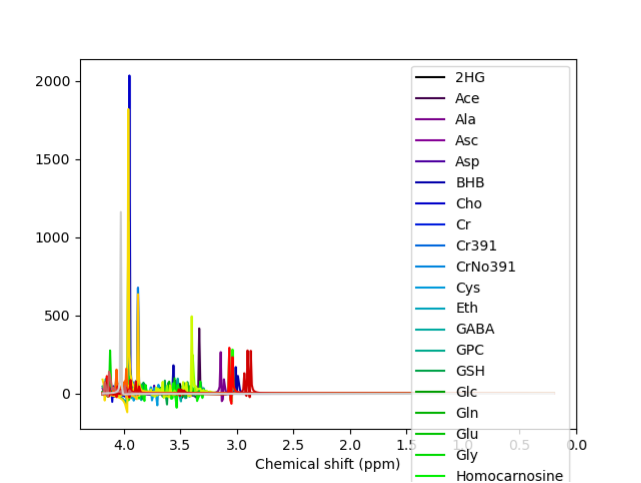
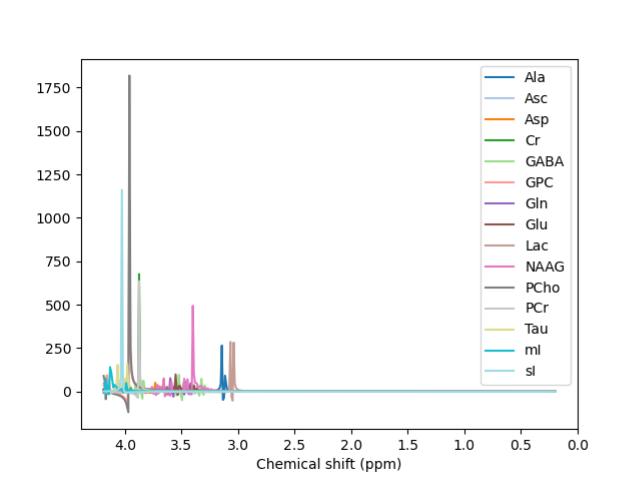
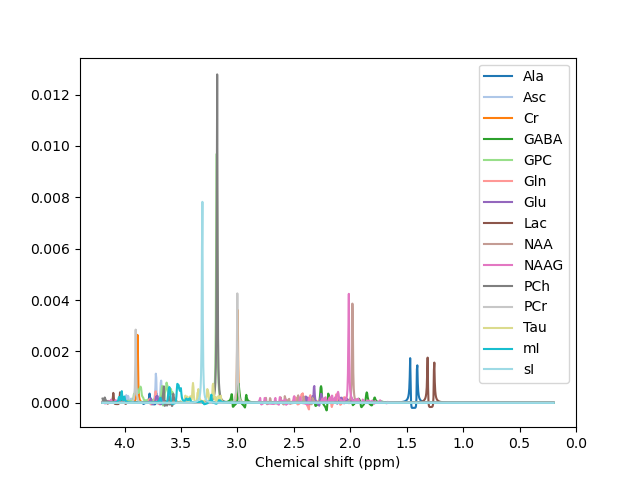
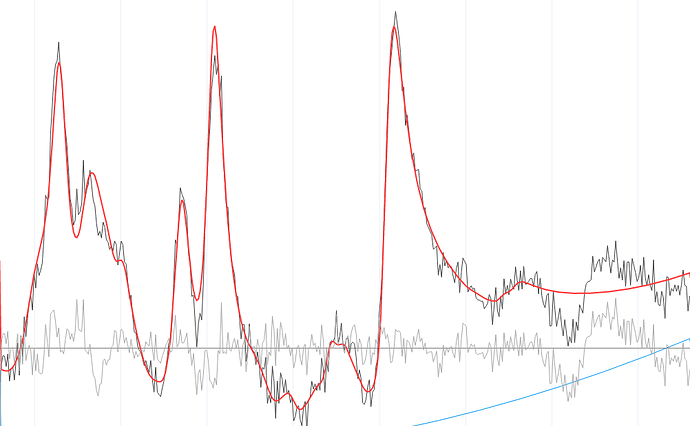
@Helge We have been able to fit the spectrum using LCModel, and the results looked much nicer. We were planning on using it anyway to validate our results from FSL-MRS. We haven’t tried using any other basis sets, so thank you for those links; we will try them and let you know if we succeed.
@alex Thank you for your help. After converting the basis set with the provided command and running the fsl_mrs command we get the following error messages:
fsl_mrs.utils.misc.InsufficentTimeCoverageError: Input data covers less time than is requested by interpolation. Change interpolation points or dwell time.
fsl_mrs.core.basis.BasisHasInsufficentCoverage: The basis spectra covers too little time. Please reduce the dwelltime, number of points or pad this basis.
Could there be an issue with the data itself? Comparing the info for the data and basis we get:
NIfTI-MRS version 0.5
Data shape (1, 1, 1, 1023)
Dimension tags: [None, None, None]
Spectrometer Frequency: 123.261637 MHz
Dwelltime (Bandwidth): 8.334E-04s (1200 Hz)
Nucleus: 1H
Field Strength: 2.90 T
------- Basis Object ---------
BASIS.NMetabs = 34
BASIS.timepoints = 1024
BASIS.centralFreq (MHz) = 127.728
BASIS.bandwidth (Hz) = 2500.0
BASIS.dwelltime (s) = 4.00000e-04
Metabolites: [‘2HG’, ‘Ace’, ‘Ala’, ‘Asc’, ‘Asp’, ‘BHB’, ‘Cho’, ‘Cr’, ‘Cr391’, ‘CrNo391’, ‘Cys’, ‘Eth’, ‘GABA’, ‘GPC’, ‘GSH’, ‘Glc’, ‘Gln’, ‘Glu’, ‘Gly’, ‘Homocarnosine’, ‘Lac’, ‘NAA’, ‘NAAG’, ‘PA’, ‘PCho’, ‘PCr’, ‘PE’, ‘Tau’, ‘Thr’, ‘Try’, ‘Tyr’, ‘Val’, ‘mI’, ‘sI’]
The bandwidth * dwelltime is 1.00008 and 1, respectively. Could this incongruency lead to the errors we see?
Kind regards,
Thomas Cawood