Hi everyone,
I recently started my MRS analysis and, after some hurdles, managed to obtain some results (more details in my previous post: Error in FSL-MRS Quantification: “Specified metabolite isn’t in the list of basis spectra”)
Acquisition Details
Sequence: MEGA-PRESS
TR: 3000 ms
TE: 68.00 ms
ON editing pulse: 1.90 ppm
OFF editing pulse: 7.50 ppm
Issue: NAA Peak Reduction During Preprocessing
I noticed that my NAA peak was significantly reduced during preprocessing, and I wasn’t sure why.
Upon closer inspection of my data structure, I found the likely cause:
- Each acquisition contains 32 ON and 32 OFF transients, and there are 4 acquisitions in total.
- The Siemens 3T scanner automatically combined these into two (DICOM) files, that I convert to NIFTI after.
- However, I have no clear indication of which file corresponds to ON and which to OFF sequences.
Visualization and Processing Attempts
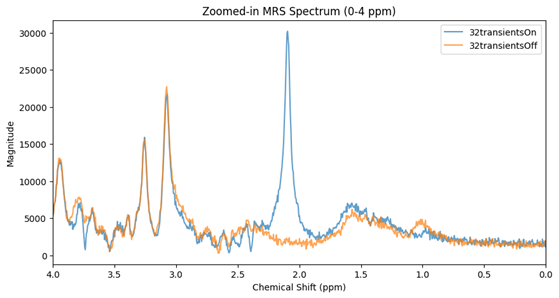
When visualizing the two original files, I get the following:
When I processed the data using FSL-MRS preproc, it automatically averaged the two files. However, I have no clear indication of which file corresponds to the ON sequence and which to the OFF sequence.
To address this, I separated them and processed them individually. This allowed me to compute the difference spectrum (OFF - ON), which resulted in either normal or inverted NAA peak (see data structure ppt)
Current Problem: Basis Set & Fitting Issues
Now, when I try to fit my difference spectra using the difference basis set that I generated on MRIcloud.org, I encounter the following error:
QuantificationError: Metabolite reference [‘Cr’, ‘PCr’] has not been fit (conc=0). Please choose another or refine fit first.
I have attempted several troubleshooting steps, but none have resolved the issue:
- Removing H₂O from the fitting sequence, but the resulting fit was very messy.
- Preprocessing the ON and OFF sequences separately and then subtracting them, but the issue persisted.
Request for Assistance
Does anyone know how I can generate an appropriate difference basis set for this specific scenario?
Are there alternative approaches I should consider?
Here is the fitting code I am currently using:
fsl_mrs --data fsl_mrs_proc/metab.nii.gz \
–LCModel_Siemens_MEGA_PRESS_Glu68_diff.BASIS \
–output fsl_mrs_fit \
–h2o fsl_mrs_proc/wref.nii.gz \
–TE 68 \
–report
Also, I am not sure if it’s okay to exclude --metab_groups (MM09 MM12 MM14 MM17 MM21), as adding them results in errors. I assume this is because they are not present in my basis set, but I would appreciate some input.
Additionally, I would love feedback on whether my analysis approach seems methodologically sound.
Thank you so much for your help
Below is an example of my full data structure:
data structure.pptx (618.7 KB)